
Skin-omics: exploring the volatile organic compounds on human skin
Nita McKenzie, Student, Liggins Institute; Dr Chris Pook, Research Technician, Auckland University of Technology; Dr Elizabeth McKenzie, Postdoctoral Fellow, Liggins Institute (Project Leader)
Overview
Skin is the largest organ of the human body. Its importance in health has only just begun to be investigated. There are hundreds of compounds on the skin surface, and they have the potential to answer many questions. These compounds can be used for diagnosis, monitoring pollutant exposure, and for understanding the skin microbiome. The ease with which skin can be sampled and analysed makes it an attractive choice for researchers. However, the major drawback of skin sampling is sample stability. Volatile samples quickly evaporate if left at room temperature, or stored in inappropriate containers. A method for stable bio-banking of human skin surface samples could open a new world of possibilities for the study of human health and the environment.
Effect of storage on the metabolite profile of human skin surface samples
The project tested the feasibility of long term storage of skin samples using miniature silicone-coated glass magnetic laboratory stir bars. Samples were collected by rolling the stir bars on the forearm. Half of the stir bars were analysed immediately using thermal desorption gas chromatography-mass spectrometry (GC-MS). The other half were stored for one month at -80°C in glass vials, then were analysed in the same manner.

Figures above: Student Nita McKenzie and Research Technician Dr Chris Pook.
Skin sampling, storage and analysis.
We discovered 330 organic compounds including fatty acids, alkanes, alkenes, azoles, amines, sulfides, aldehydes, acetates, ketones, and terpenes. We also found many compounds, including plasticisers that originated from environmental pollution and personal care products. The majority (86%) of the compounds we discovered remained stable after one month storage, showing that the integrity of the metabolic profile was not significantly compromised by storage. Storage of skin samples for long periods therefore appears feasible under the right conditions. This was the first project that trialled untargeted discovery metabolomics data extraction from start to finish on the metabolomics virtual machine, set up for metabolomics data extraction by the Centre for eResearch.
Previously, analysis of high-dimensional omics data of this type involved installing specialist software on a computer for the student, liaising with the ITS to find a computer powerful enough to process the data, or the student had to book to use the single computer available at the metabolomics instrument laboratory. The virtual machine enabled the summer student to access the data processing environment she needed from a standard university’s computer. This represented a significant saving in setup cost and time, which allowed us to focucs more time on the science.
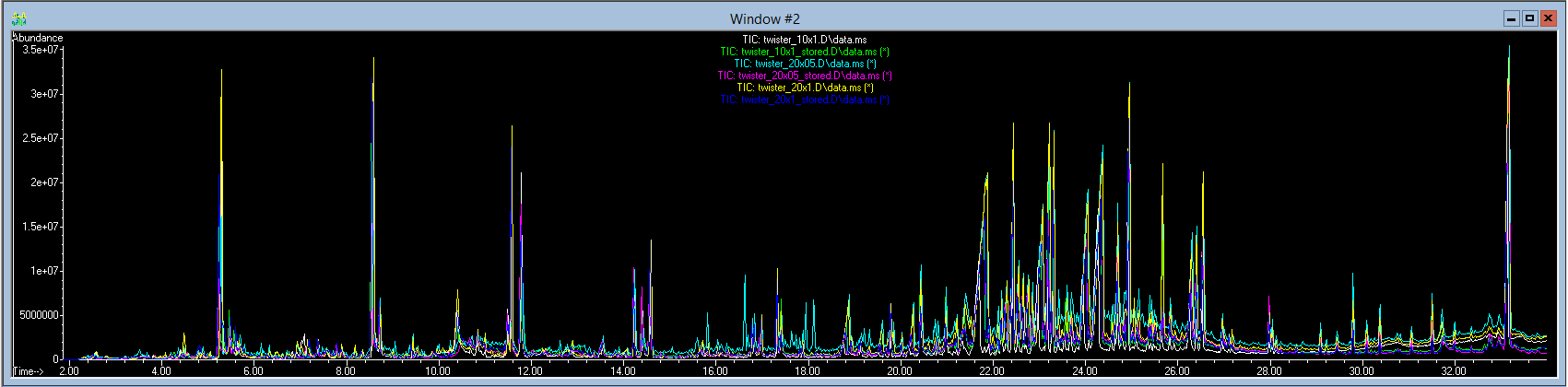
Figure 1. Overlaid chromatographic profiles showing compounds (peaks) detected from human skin samples.
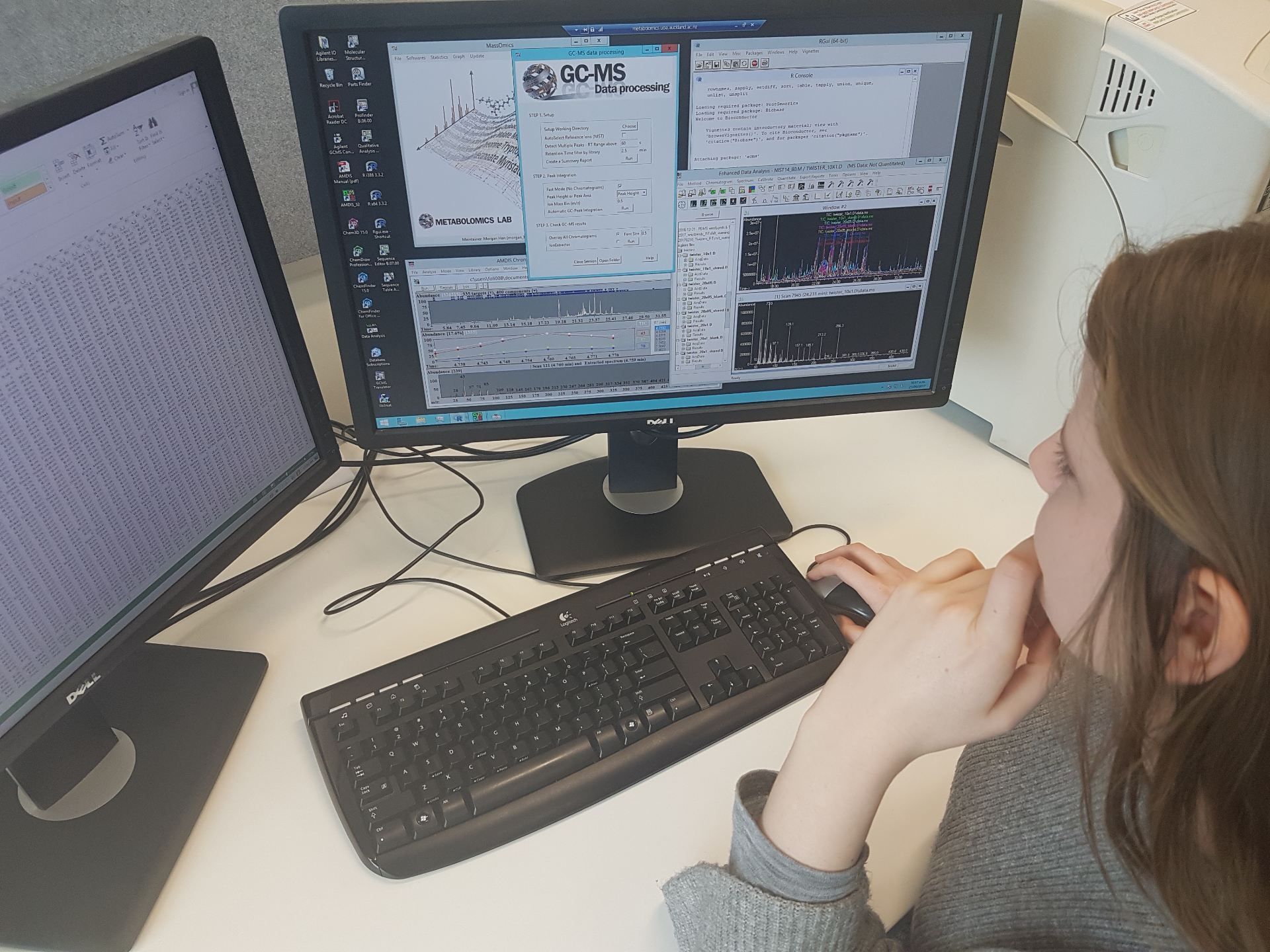
Figure 2. Data extraction and pre-processing by Nita McKenzie using the metabolomics virtual machine.
The future of the project
The purpose of this study was to assess the feasibility of using human skin samples for detection of biomarkers of acute illness from infant skin in neonatal intensive care. The results from this study were presented to the Australia and New Zealand Society for Mass Spectrometry Conference in July 2017 and the data is now being used to support grant applications for research on biomarkers of acute illness from infant skin.
This collaboration with the Centre for eResearch not only advanced science in the field of human skin diagnostics, it also provided support for the ideas and research of early career researchers from both the Liggins Institute and Auckland University of Technology.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































