
Myocardial motion tracking and strain calculation using deep learning networks
Edward Ferdian, Dr Avan Suinesiaputra and Professor Alistair Young, Department of Anatomy and Medical Imaging, Faculty of Medical and Health Sciences
Description
Cardiac Magnetic Resonance (CMR) is a non-invasive imaging technique to analyse patient heart. CMR is currently the gold standard among other imaging techniques due to its superb contrast, no radiation exposure and its versatility to produce different images in a single examination session. CMR tagging is one of the image protocols that artificially creates stripe or grid markers on the acquired images. These markers follow the deformation of the heart, allowing high resolution of cardiac motion tracking and analysis. However, full analysis of cardiac motion from CMR tagging is still time consuming and requires significant post-processing time.
In this project, we developed a machine learning algorithm to fully automatically detect the location of landmark points inside the myocardium from CMR tagging images when the heart is at full relaxation (end-diastole) and track the motion of these points in subsequent image frames throughout the whole cardiac cycle. We built two deep learning neural networks for these tasks. The first network was trained to locate a bounding box covering the myocardium and it was based on 7 layers of convolutional neural networks (CNN). The output of this network is fed into the second network, which was trained to detect landmark points and follow their movements. The second network was based on RNNCNN; a combination of CNN and Recurrent Neural Network (RNN) with Long Short-Term Memory (LSTM) units.
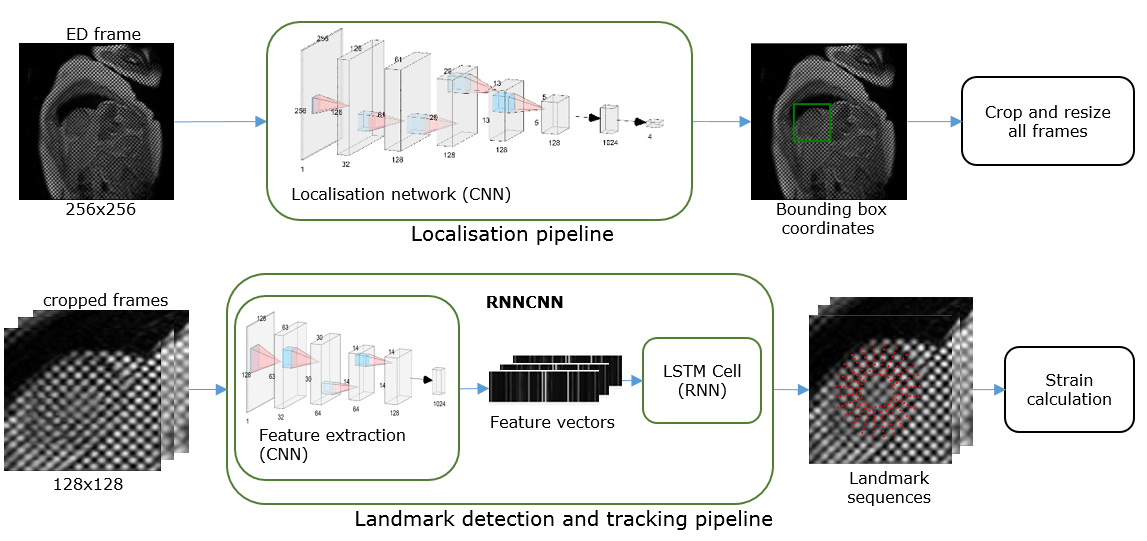
Landmark detection and tracking pipeline
Training the networks
Deep learning networks require massive size of data and high computational power during training. We trained the whole pipeline using 4,508 CMR cases, obtained from the UK Biobank study. In total, there were 12,409 image sequences, each consisted of 20 image frames, and the input for the network was 256×256 image pixels. The whole network consists of 39.4 millions of parameters to optimise. With the help and support from the Center for eResearch, we trained our networks on the Nectar Research Cloud with GPU processing capability (NVIDIA Tesla K40). The training process required approximately 12 hours using TensorFlow version 1.5.0 library for deep learning.
Results
We calculated myocardial strain from the landmark points predicted by the networks and compared with expert analyses. The predicted strains were within 1% of the manual ground truth on average, except for basal radial strain (2.4%). The predicted strains also showed comparable intraclass correlation coefficient with the manual ground truth. This fully automated analysis of CMR tagging, including strain calculation, could detect landmarks and predict strains up to 920 frames per second (~4.5 minutes for the whole 12,409 image sequences). In future, we are aiming to develop an integrated machine learning analysis for cardiac MRI, including segmentation, breath-hold mis-registration, diagnosis and prognosis of cardiovascular disease, and 4D Flow analysis without any manual interventions. Without the access of the GPU-powered Nectar cloud, this research would not be practicable.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































