
Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model
Dr Juliette Cheyne, Research Fellow; Professor Johanna Montgomery; Laura McNamara, Doctoral Candidate; Department of Physiology
Background
Alzheimer’s disease (AD) is the most common neurodegenerative disease, causing an ultimately fatal progressive and incurable cognitive decline. The key pathology of AD is the accumulation of toxic amyloid plaques throughout the temporobasal and eventually frontomedial areas of the brain (Alzheimer’s Disease International, 2018). Subtle neuronal changes occurring early in AD progression such as synapse loss and circuit changes have long been known to precede eventual widespread neurodegeneration (Davies et al., 1987). Technological limitations have restricted our ability to collect longitudinal data reflecting these dynamic circuit changes. However, the recent development of head-mounted miniaturised fluorescence microscopes (miniscopes) allows the chronic recording of neuronal circuit activity while rodents engage in active behaviours (Shuman et al., 2020).
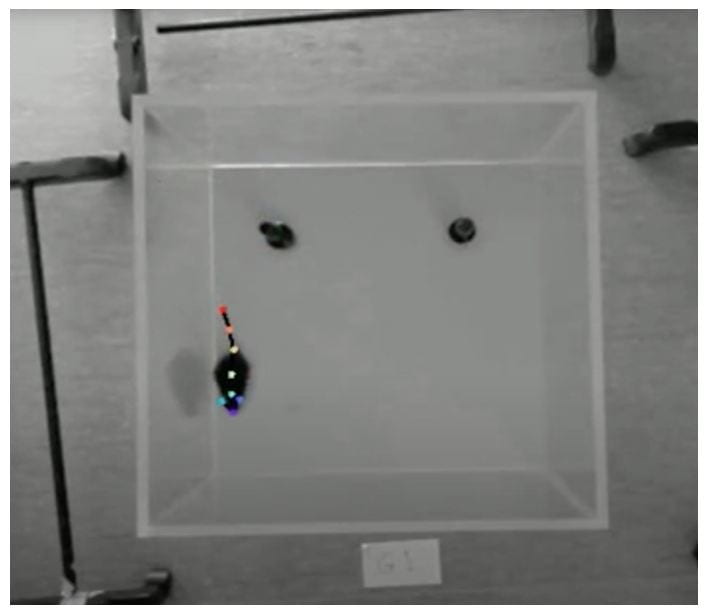
The aim of our research is to utilise miniscope technology in a murine model of AD to characterise correlations between neuronal activity in the CA1 hippocampus, behavioural deficits and amyloid-beta plaque load. To accomplish this, aged male APP/PS1 mice were hippocampally injected with a GCaMP7s viral vector to report pyramidal cell calcium activity. They were subsequently implanted with a Graded-Index lens and affixed with a baseplate for imaging and miniscope attachment, respectively. Mice underwent two spatial memory behavioural testing sessions at 15 and 23 months of age, while the attached miniscope concurrently recorded CA1 calcium activity.
To analyse the behavioural component of this data, DeepLabCut software was utilised. This machine learning software package automates obtaining positional information via training a network on a dataset of hand-labelled frames. This allows it to estimate the position of each body part of interest in frames from a similar recording (Mathis et al., 2018). These outputs are further analysed to derive information about the animal’s position, motion and engagement with its environment, which can in turn be correlated with cellular outputs.

Problem
Machine Learning tasks and DeepLabCut can be significantly sped up by using a Graphical Processing Unit (GPU). Ubuntu was selected as the Linux distribution running as a virtual machine (VM) on the Nectar Research Cloud.
As Miniscope work generates data files in the magnitude of hundreds of gigabytes, these large amounts of data had to be moved securely and efficiently to the VM. These double in size after DeepLabCut is run due to the generation of additional video output files. The inability to reliably move these files between local PCs and VM locations essentially halted our research progress.
Approach
Center for eResearch supported this research project by installing and updating the complex software package DeepLabCut. For the file transfer, the Globus Connect software was chosen. Its setup required changes to the user and access right management on the virtual machine that was efficiently and reproducibly achieved by customised scripts.
Results
The overall process was carried out in close collaboration with the researchers who gained experience in a Linux environment working with Python and becoming significantly more confident with respect to troubleshooting within an Ubuntu terminal and in Nectar. File transfers can be initiated by the researchers and take less time. Different users on the VM can see their contributions. The use of the GPU reduces the processing time of the Machine Learning algorithms significantly. Based on their learning, the researchers have since established another VM allocated specifically for running RAM-heavy MatLab scripts, which has optimised the cellular component of our research as well.
- Alzheimer’s Disease International. (2018, September 6). World Alzheimer Reports | Alzheimer’s Disease International.
https://www.alz.co.uk/research/world-report - Davies, C. A., Mann, D. M. A., Sumpter, P. Q., & Yates, P. O. (1987). A quantitative morphometric analysis of the neuronal and synaptic content of the frontal and temporal cortex in patients with Alzheimer’s disease. Journal of the Neurological Sciences, 78(2), 151–164. https://doi.org/10.1016/0022-510X(87)90057-8
- Shuman, T., Aharoni, D., Cai, D. J., Lee, C. R., Chavlis, S., Page-Harley, L., Vetere, L. M., Feng, Y., Yang, C. Y., Mollinedo-Gajate, I., Chen, L., Pennington, Z. T., Taxidis, J., Flores, S. E., Cheng, K., Javaherian, M., Kaba, C. C., Rao, N., La-Vu, M., … Golshani, P. (2020). Breakdown of spatial coding and interneuron synchronization in epileptic mice. Nature Neuroscience. https://doi.org/10.1038/s41593-019-0559-0
- Mathis, A., Mamidanna, P., Cury, K. M., Abe, T., Murthy, V. N., Mathis, M. W., & Bethge, M. (2018). DeepLabCut: Markerless pose estimation of user-defined body parts with deep learning. Nature Neuroscience, 21(9), 1281–1289. https://doi.org/10.1038/s41593-018-0209-y
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































