
Improving arrival time predictions for vehicles in a public transport network
Tom Elliott, Doctoral Candidate, Department of Statistics
Predicting the arrival time of buses
In recent years, vehicle tracking technologies (GPS) have advanced along with communication technologies (internet, mobile apps) to the point where public transport users can see the real-time location of their buses on a map through their phones. However, the information on estimated arrival time (ETA) are still undependable. In place of statistically-founded predictive models that have been used historically, ETAs are often calculated based solely on the vehicle’s current delay, as is the case here in Auckland. These predictions assume that the timetables are accurate (they often are not), and that the delays will not be affected by traffic conditions (they often are). Thus, the predictions are frequently inaccurate and ultimately unreliable.
Modelling network status using real-time data
We have been working on a predictive modeling framework that uses real-time vehicle locations to estimate network state (i.e., bus speeds along roads within the network), which is then used to generate ETAs that reflect current traffic conditions. This involves three stages:
- a vehicle model that uses vehicle location data, freely available via Auckland Transport’s public API, to estimate the vehicle’s state: how far (in meters) it has traveled along the route, its speed, and travel times along the way.
- a network state model that takes the estimated travel times of all vehicles, irrespective of route, and models the current average travel time of transit vehicles traveling along each road in the network; and
- an arrival time prediction model that combines the estimated vehicle state from (1) with estimated travel times from (2) to predict the arrival time at all upcoming stops, taking into account uncertainty due to traffic lights and intermediate stops. (see Figure 1)
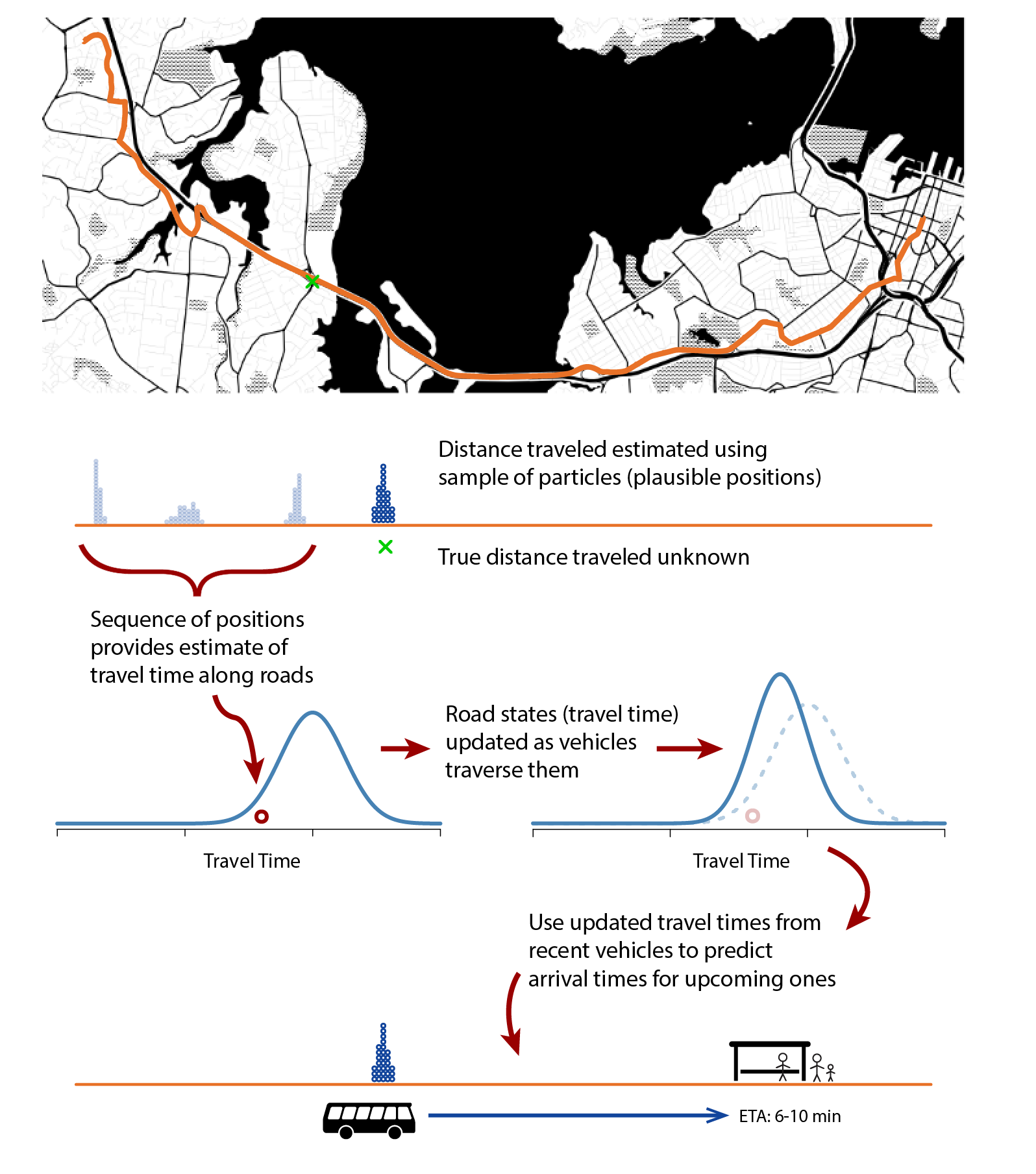
Figure 1. Real-time vehicle locations are used to model travel times at are used to predict arrival time.

Figure 2. The VM processes real-time positions of all Auckland buses to provide ETAs to commuters.
How the virtual machines make it possible
We are using a particle filter to implement the vehicle model, as it provides a robust, extensible framework; in particular, it handles multimodality (for example at bus stops), and allows us to use an intuitive likelihood function (based on the distance between two points on a map).
The downside of this particle filter is its high computational demand: at peak times, there are upwards of 700 buses operating, each modelled using 1000’s of particles. These particles are also used in arrival time prediction, so predictions are required for every upcoming stops for all particles of each bus. In addition, this all needs to run efficiently in real-time to ensure ETAs are available to commuters as soon as possible, not 5 minutes after receiving the data.
A dedicated virtual machine is the ideal tool for the job. It can continually update ETAs in real-time as new data is received, and be supplied with enough resources so that the entire model runs in under 30 seconds. After contacting staff at the Centre for eResearch, they quickly assessed our needs and had a RVM up and running for our use. (see Figure 2)
Future work
Our goals for the future are to have the full model running on the VM in real-time, generating arrival time predictions for all future stops of active trips. We then hope to investigate using historical data, along with real-time, to estimate delays for trips that are yet to begin (currently, these are assumed on-time until the bus begins the trip). We will also look into journey planning applications that make use of our results and deciding on the “best route” to take: for example, which route is the fastest, or most likely to get me to work on-time.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































