
3D Cryo-EM reconstructions of macromolecular complexes
Hariprasad Venugopal, School of Biological Sciences
To understand biology, it is key to understand the structure of bio-molecules.
Late 20th century has yielded great advancements in techniques like X-ray crystallography and Nuclear Magnetic Resonance (NMR) to delve into the structures of biomolecules to atomic resolution as well as to understand its internal dynamics. Cryo–electron-microscopy (Cryo-EM) is a recent and fast developing tool to directly visualize these structures. The study of large biomolecular assemblies, which are generally intractable by X-ray and NMR has been opened up to near-atomic resolution to atomic resolution with the recent advances in Cryo-EM imaging and image processing. With the procurement of the TF20 transmission electron microscope enabled with energy filtering and tomographic capabilities at the University of Auckland, this kind of work has become feasible for the first time in New Zealand Pan cluster provides the necessary computational platform for processing high resolution images which are computationally very expensive.
Armed to deliver – Insight into the action of a microinjection nanodevice
This is a Marsden funded project for elucidating the functional mechanism of a microinjection device, AFP (anti-feeding Prophage), a complex of 18 different protein, evolved by bacteria Serratia entomopihila that delivers a toxin to kill Cosselyira zealandica, commonly known as grass grub, a widespread pest of pasture that causes huge economic loss to New Zealand.
This work is in collaboration with Dr. Mark Hurst of AgResearch. In a recent paper titled Three-dimensional Structure of the Toxin-delivery Particle Antifeeding Prophage of Serratia entomophila (Heymann, J. B., J. D. Bartho, et al. (2013). JBC 288(35): 25276-25284), we have elucidated the structure of the protein complex in resting state to a resolution of ~20Å, which shows the domain level architecture of the nano-device as well as its toxin cargo encased in the central sheath. With high resolution data from the new microscope we will be pushing towards capturing the particle in its various conformational snapshots leading all the way to the contracted state at atomic resolution.
This detailed understanding of such a mobile protein delivery system and its unusual eukaryotic target specificity could guide rational designing of nanoscale devices and hence opening cutting edge therapeutic avenues such as delivering antigenic proteins in anti-tumour immunotherapy.
Why Pan cluster?
3D reconstructions from 2D EM images of biomolecules are performed from thousands of images of the molecule. This kind of image processing is computationally very expensive. Modern Cryo-EM image processing software like BSOFT and EMAN (1, 2.1) acknowledges this issue and is written to incorporate scalability in terms of multi thread processing abilities. Pan cluster offers an ideal platform to make use of the parallelisation in these computational routines to fasten the calculation. A single particle analysis routine consists of aligning the particles picked from the micrographs to a reference model. This reference is refined in an iterative way to reach final 3D reconstruction. For example in BSOFT, such an alignment of Afp particle defined in a 400*400 pixel image at 42k magnification scanned at 3.02 Å/pixel raster-step, takes 180 seconds per particle on our workstation (Intel® Core™ i7 CPU 960 @ 3.20GHz processor with 8 threads and 8GB RAM). This single particle orientation and alignment searches are linearly scaled, i.e. 1 processor per particle. When the datasets consists of 105-106 particles (critical for atomic resolution studies) the computational time required on a standard PC will be in the order of months. Pan offers better hardware and large number of multi-core-processors which are set up for parallel computation. Present calculations use 10 processors on a single node but for higher number of particles as mentioned earlier this can be scaled to use multiple nodes. Such an infrastructure enables us to run multiple jobs with large number of processors in order to perform various computational tests in a timely manner hence making this endeavour feasible.
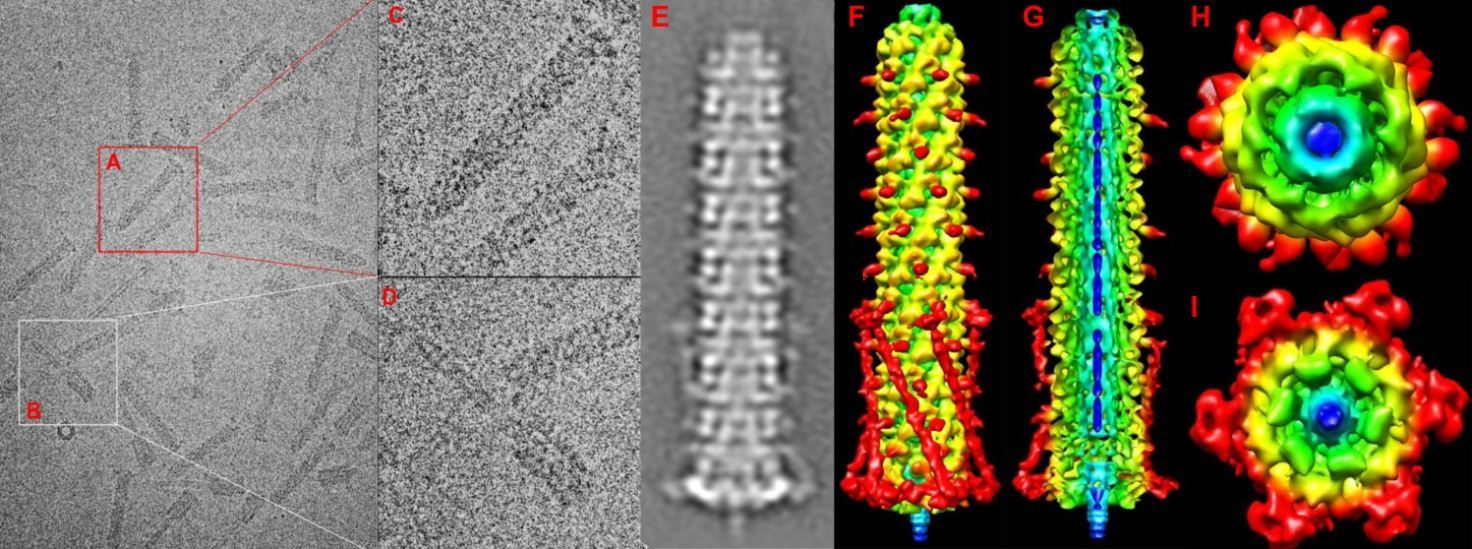
Figure 1: A: 400*400 pixel boxed AFP particle B: Boxed AFP contracted form. C: Enlarged view of Afp Particle D: enlarged view of the contracted particle. E: Stacked image of Afp particle of similar view. F: Radially coloured 3D reconstruction of Afp particle from blue (center) to red (periphery). G: Vertical cross section of the Afp particle showing the central cyan tube containing the putative toxin colured blue. H: Shows the top view of the Afp particle with sheath proten exhibiting (orange coloured) helical rotation. I: Bottom view of the base plate with the blue puncturing tip.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































