
3D Electromagnetic modeling and simulation using heterogeneous computing
John Rugis, Institute of Earth Science and Engineering.
In the earth sciences, a number of different electromagnetic techniques are used to non-invasively acquire information about underground material properties and objects.
One of these techniques, magnetotellurics, takes advantage of the naturally occurring time varying electromagnetic field that surrounds the earth. The lowest frequency components of this field penetrate through the surface of the earth to a depth of up to tens of kilometres. Differing subsurface materials reflect this field and careful measurements of these reflections can be used to create 3D images of the subsurface through a computational process known as inversion.
The goal in this project was to explore the possibility of speeding-up the most time consuming step of the inversion process – the forward modelling. To achieve this goal, we selected one of the highest performance options available in computing today: Intel CPU’s and NVIDIA GPU’s across multiple nodes.
Simultaneous utilisation of multiple compute nodes requires careful problem decomposition and parallel programming techniques. However, before taking on this complexity, it is generally best to start with a reference model. Results produced by subsequent parallelisation can then be verified against the reference. Figure 1 shows a reference visualisation of simulated electric field in a 100 × 100 × 100 model space after 120 time steps. The results after parallelisation and scaling-up were a perfect match!
The final model size in this project was increased to 848 × 848 × 848. A slice of the electric field after 550 steps is shown in Figure 2 where we see the concentric rings which is exactly what electromagnetic theory says should happen.

Figure 1: Electric field in reference model
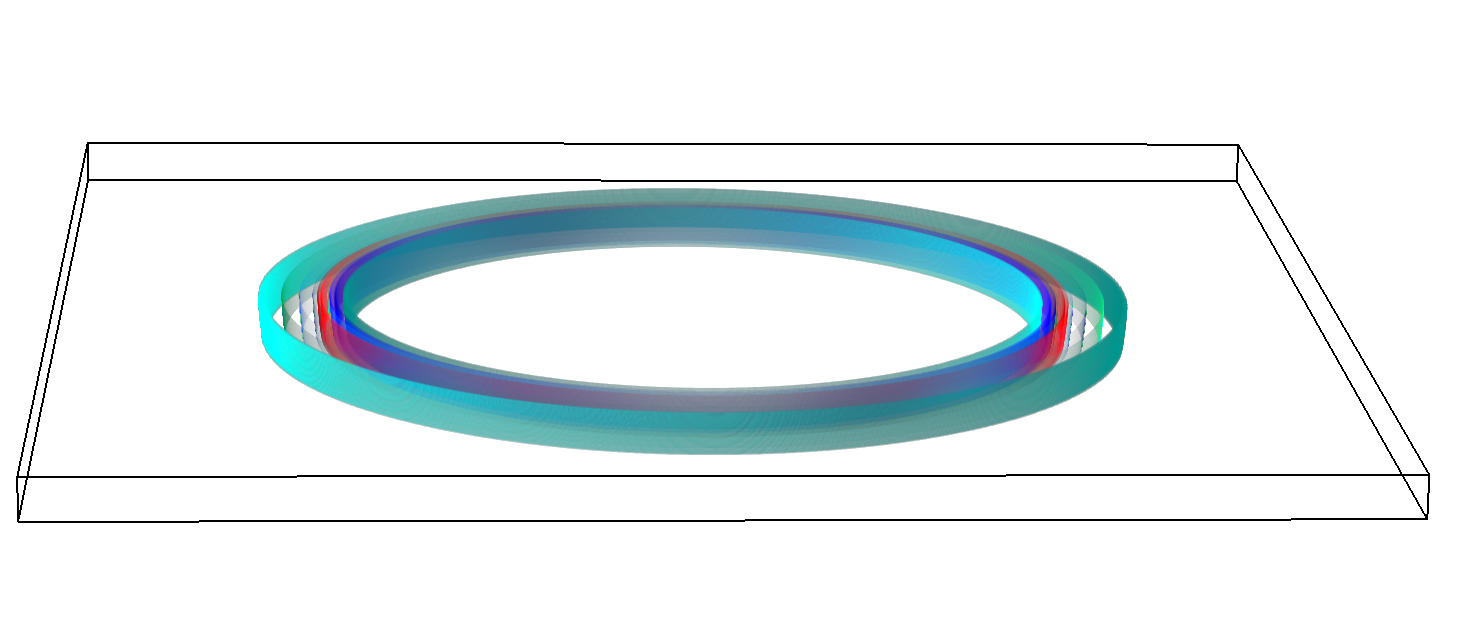
Figure 2: Electric field slice in final model
Using GPU nodes
Using eight GPU nodes on the Pan cluster at the University of Auckland, we were able to achieve an 80-fold overall speedup in forward modelling run time. With this success, we are on-track to reduce typical 3D magnetotelluric inversion run times from over a week to less than a day. The NVIDIA CUDA software development tools on the NeSI Pan were used extensively for coding, debugging and profiling. OpenMPI libraries were used for inter-node communication. We could not have even considered this work without access to specific high performance computing facilities such as those available at the University of Auckland. The support and technical assistance from the staff at the Centre for eResearch of the University of Auckland was instrumental in our success. Parallel computing is central in our research leading to more robust and efficient methods. Our parallel computing success has helped us to meet and exceed client expectations in our commercial work. We plan to further optimise our techniques and to continue scaling up to use additional GPU nodes as they become available on the NeSI Pan cluster.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool























































































































































