
Accelerating the discovery of natural products made by orphan megasynthases
Verne Lee, School of Biological Science
Natural products and orphan megasynthases
Natural products are bioactive molecules produced by living organisms. These molecules play important biological roles in signalling, nutrient acquisition and defence. They are also very important compounds in human and animal health as natural (often foodborne) toxins and particularly as sources of novel pharmaceuticals. Some of the most chemically diverse and useful natural products are made by microbes using a specialised class of enzymes known as “megasynthase” enzymes (see Figure 1).
Large-scale sequencing of microbial genomes has revealed a large number of genes that code for megasynthase enzymes. However, for a big subset of these megasynthases nothing is known about the natural products that they produce. These megasynthase are often referred to as “orphan” megasynthases. The production of natural products by microbes is typically under very tight control and they are only produced in specific conditions. This makes identifying and studying the natural products made by orphan megasynthases very challenging.
Megasynthase enzymes function in a manner analogous to an assembly line, joining together multiple chemical building blocks one after another to produce the finished natural product. Each unique natural product is made by a unique megasynthase enzyme. The chemical building blocks that will be used to assemble the natural product are selected by specific parts of the megasynthase enzymes. These parts of the megasynthases can be referred to as “binding domains” as they select the specific building blocks from the pool available in the cell by specifically binding them in a binding pocket. Figure 2 shows the binding pocket of a binding domain with a building block bound into it.
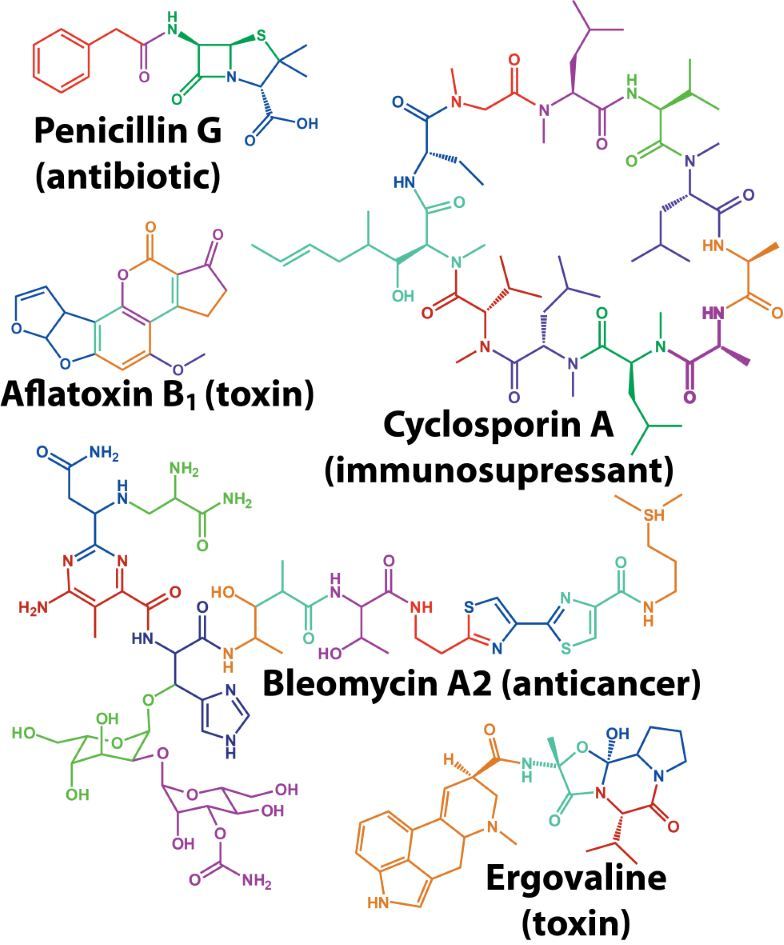
Figure 1: Megasynthase enzymes are capable of producing a diverse range of natural products. A few examples of natural products produced by megasynthase enzymes are shown. Megasynthases assemble the natural products from multiple chemical building blocks, depicted here in different colours.
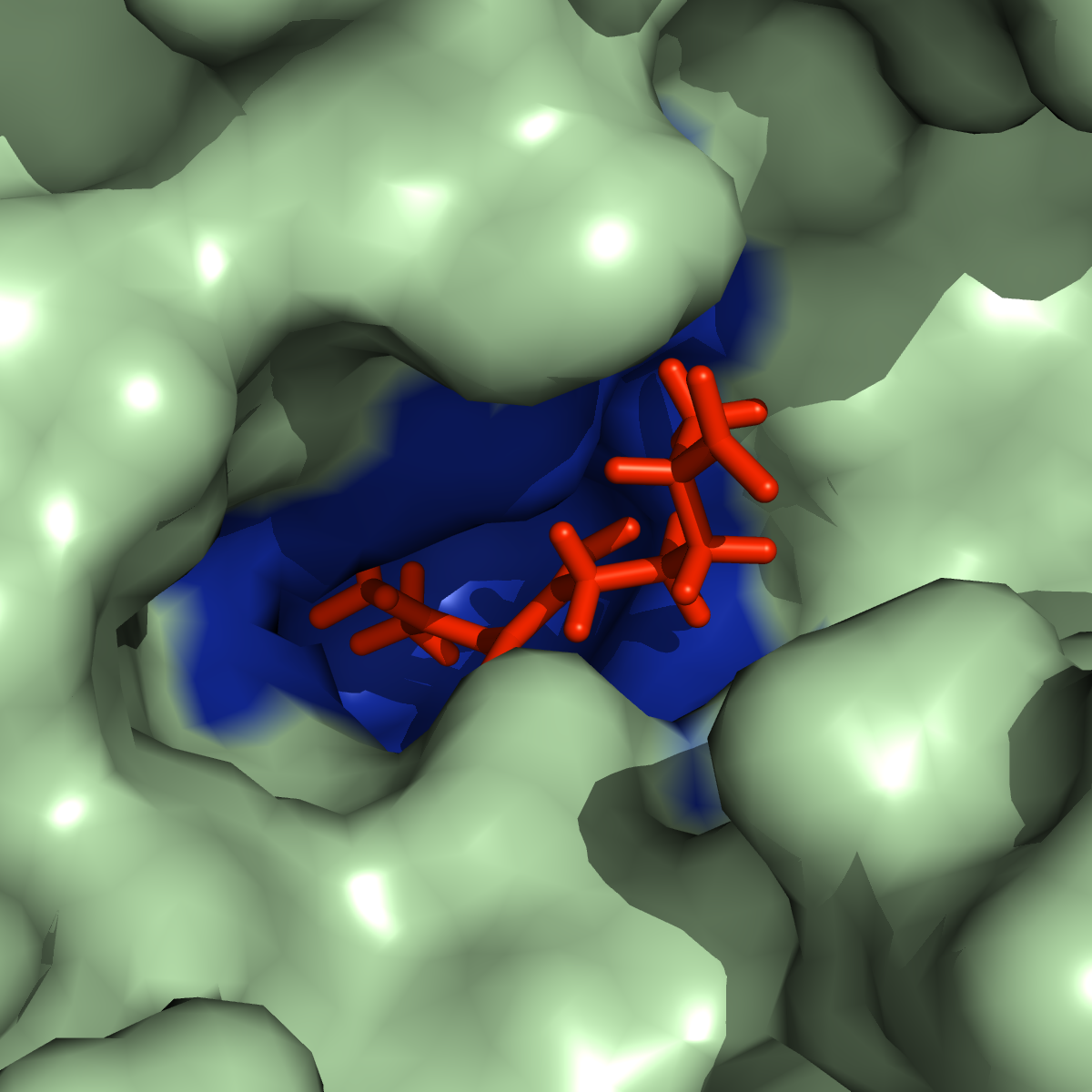
Figure 2 (above): The chemical building blocks used to assemble the natural products are selected by binding in the binding pockets of the megasynthase binding domains. A chemical building pocket (red) is depicted bound into the binding pocket (blue) of a binding domain (green).
Predicting the products of orphan megasynthases
If we were able to predict the characteristics of the natural product that a megasynthase produces based on the gene sequence of the orphan megasynthase, this would go a long way towards identifying and investigating these novel natural products. A key part of this effort is predicting which chemical building blocks are used by a megasynthases to assemble its natural product. As the building blocks are selected by the specific binding pocket in the binding domain, this information is available in the gene sequence. However, the predictions cannot be made directly from sequences. Instead, a two-step process is required. The shape and nature of the binding domain and respective binding pocket must be modelled computationally. Then, the process of binding between potential building blocks and the modelled binding pocket must then be simulated to predict which building block is bound tightest by the binding domain.
Simulating binding
Simulating the binding of the potential building blocks to the modelled binding domains is particularly challenging. Our technique involves simulating the forces involved in the interactions between the atoms of the binding domain, the potential building block molecules and surrounding water molecules using the Amber molecular dynamics package on the Pan cluster.
The system simulated can include over 50,000 atoms that are in constant motion (Figure 3). Furthermore, these simulations must be done accurately for a fairly large number of potential building blocks (we have been conducting trials with 160 potential building blocks) within a reasonable time frame. With the binding simulations on each building block able to be run independently, parallel computation on the Pan cluster was ideal for our needs. Together with the parallelisation capabilities of the Amber package, which allowed each simulation to be spread over multiple cores, the run time for a full set of 160 building blocks is on the order of 3-4 days. Without the Pan cluster, our simulations are simply not feasible.
International collaboration – putting it together
We are conducting our research in collaboration with researchers at Aarhus University, Denmark, who are developing computation techniques to predict how a megasynthase assembles the building blocks into the final natural product. The aim is to combine our predictions of the chemical building blocks with their predictions of natural product assembly to result in complete prediction of the natural products made by the orphan megasynthases. This research and collaboration is only possible with the computing resources provided by the NeSI and the excellent support from the staff at the Centre of eResearch.
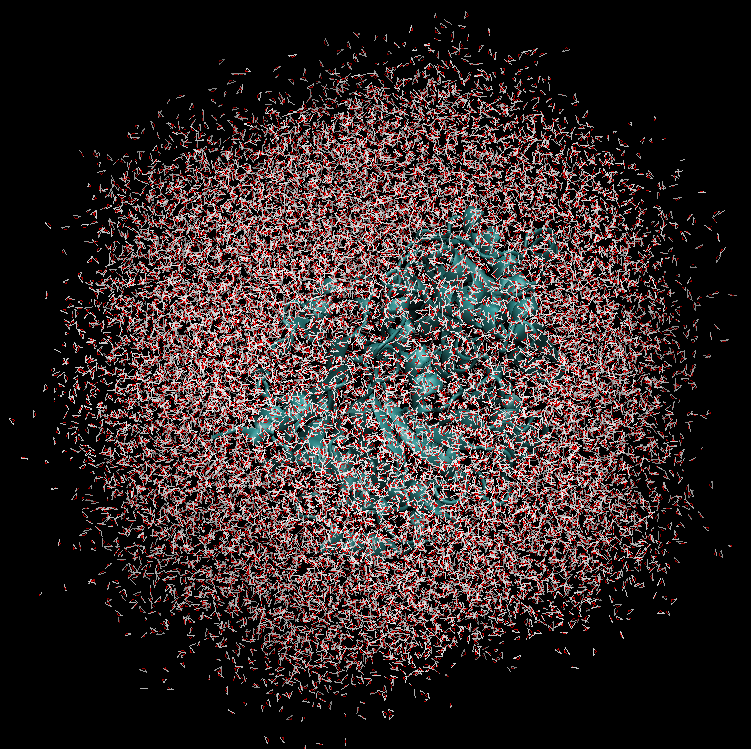
Figure 3 (right): In this snapshot from a binding simulation, a binding domain (cyan) can be seen surrounded by water molecules (white and red). The chemical building block being tested for binding is present in the simulation but is not visible in this figure.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































