
BEAST, Bayesian evolutionary analysis sampling trees
Alexei Drummond, Bioinformatics, Department of Computer Science
BEAST is a cross-platform program for Bayesian MCMC analysis of molecular sequences. It is entirely oriented towards rooted, time- measured phylogenies inferred using strict or relaxed molecular clock models. It can be used as a method of reconstructing phylogenies but is also a framework for testing evolutionary hypotheses without conditioning on a single tree topology. BEAST uses MCMC to explore tree space in such a way that each tree is weighted proportional to its posterior probability. We include a simple to use user-interface program for setting up standard analyses and a suit of programs for analysing the results.
BEAST 2 (www.beast2.org) develops new features to allow resuming an MCMC chain, and real time tracking of ESSs while running a chain.
It is easily extendible, for example, the BEAST 2 packages supports multi-chain MCMC, some experimental likelihood calculations that are potentially faster than the base implementation, and a spread sheet GUI for manipulating models.
BEAST 2 has a plug-in facility. BEAST 2 packages provide more models, such as
- Birth Death Serial Skyline Model
- Reversible jump substitution model
- SNAPP Species trees from SNP and AFLP analysis
- Substitution Bayesian Model Averaging
- Sampled Ancestors
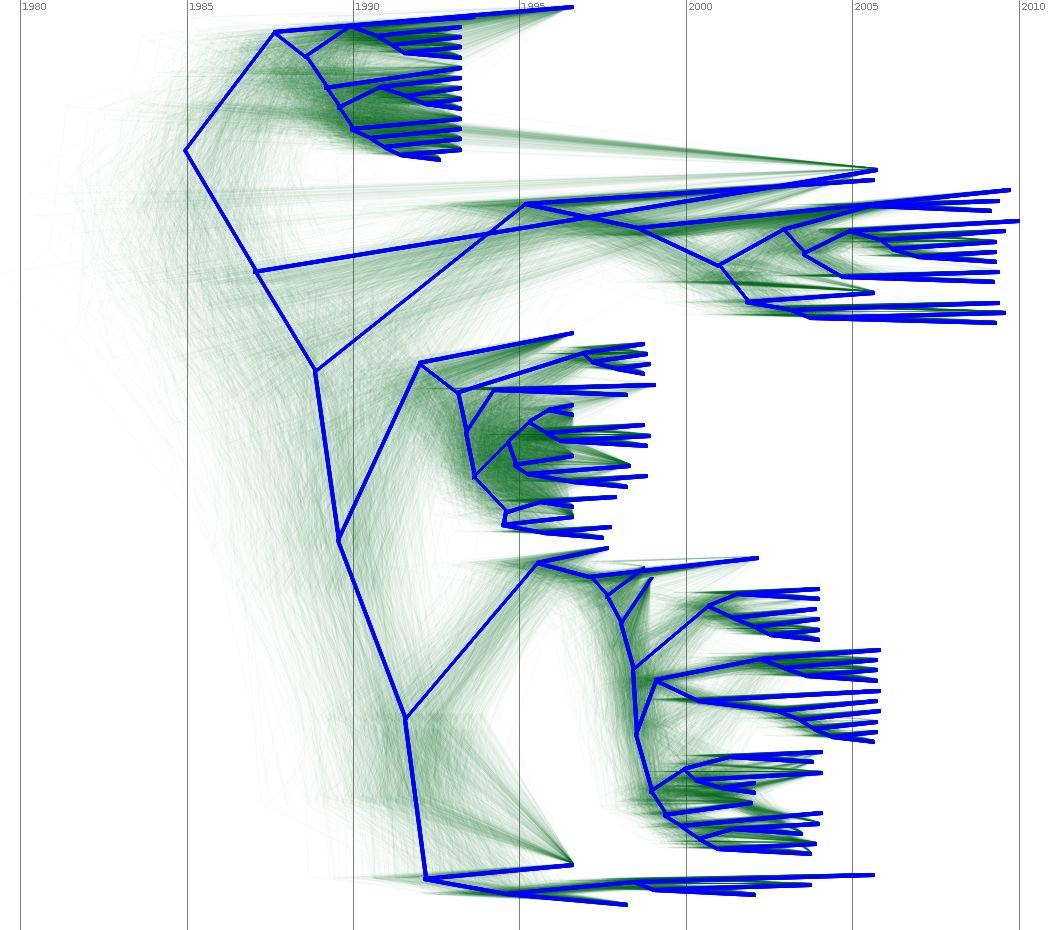
Output of a BEAST analysis of viral samples taken over different years — samples from the same year tend to cluster together. Blue line represents a summary tree of the posterior sample. Green lines indicate uncertainty in the tree distribution. It illustrates how both topology and node height estimates vary in the posterior sample.
They can be easily installed/un-installed from BEAUti (BEAST’s GUI). More packages are being developed (http://www.beast2.org/beagle-beast-2-in-cluster) .
BEAST developers use the NeSI Pan cluster for running simulation studies to validate new models, and run large analyses with questions of considerable scientific interest. For example, BEAST was used for dating the split of Cichlid species between Africa and South America, determining the origin of Indo-European languages (see http://language.cs.auckland.ac.nz), and analysing epidemiological histories of diseases such as HIV and influenza.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































