
Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis
Shuqi E. Wang, Anna E. S. Brooks, Anthony M. Poole & Augusto Simoes-Barbosa, School of Biological Sciences
Background
Trichomonas vaginalis, the causative agent of a prevalent urogenital infection in humans, is an evolutionarily divergent protozoan. Protein-coding genes in T. vaginalis are largely controlled by two core promoter elements, producing mRNAs with short 5′ UTRs. The specific mechanisms adopted by T. vaginalis to fine-tune the translation efficiency (TE) of mRNAs remain largely unknown.
Results
Using both computational and experimental approaches, this study investigated two key factors influencing TE in T. vaginalis: codon usage and mRNA secondary structure. Statistical dependence between TE and codon adaptation index (CAI) highlighted the impact of codon usage on mRNA translation in T. vaginalis. A genome-wide interrogation revealed that low structural complexity at the 5′ end of mRNA followed closely by a highly structured downstream region correlates with TE variation in this organism.
To validate these findings, a synthetic library of 15 synonymous iLOV genes was created, representing five mRNA folding profiles and three codon usage profiles. Fluorescence signals produced by the expression of these synonymous iLOV genes in T. vaginalis were consistent with and validated our in silico predictions.
Conclusions
Using complementary in silico and in vivo approaches, this study systematically investigated the impact of codon usage bias and mRNA secondary structure on TE of T. vaginalis mRNAs. In addition to guiding optimal gene expression in this parasite, the results here provide new information that leads to a more comprehensive understanding of the mechanisms by which gene expression is controlled in T. vaginalis. Despite having a translation machinery that is rather typical of eukaryotes, T. vaginalis and other evolutionarily divergent protozoans such as Giardia produce mRNAs with short 5′ UTRs. This study indicates that T. vaginalis mRNAs contain information at the level of both sequence and structure that impact their expression, adding a novel layer to our understanding of gene regulation in this divergent unicellular eukaryote.
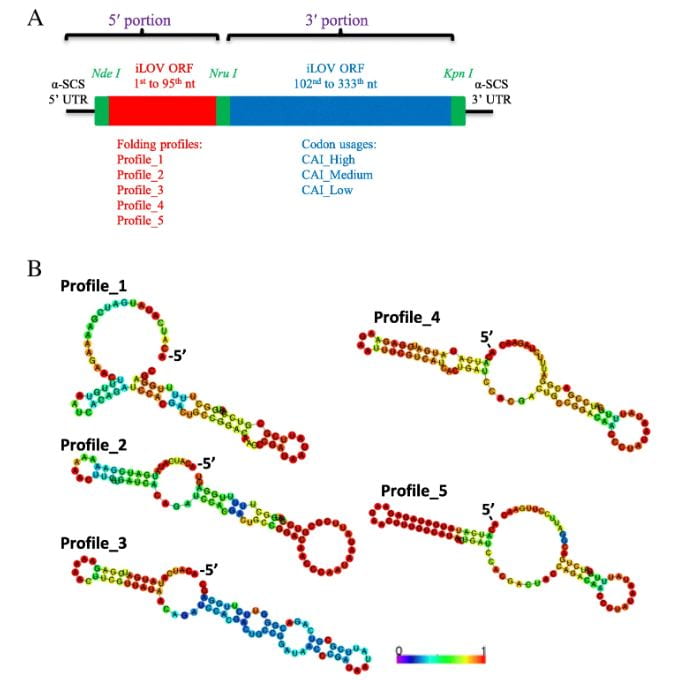
Details of the synthetic iLOV gene library to be expressed in T. vaginalis.a Diagram of the iLOV genes showing 5′ and 3′ portions connected by the Nru I site.
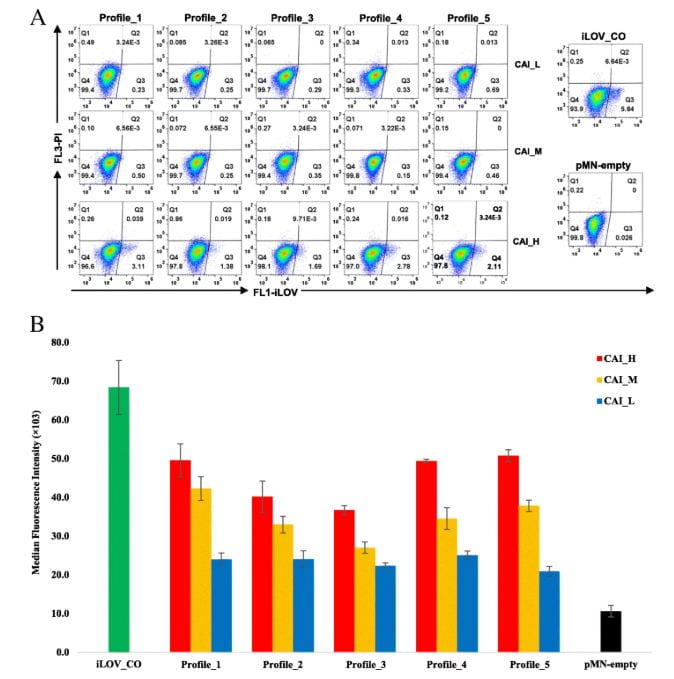
Transient and stable expressions of the synonymous iLOV genes determined by flow cytometry.
Acknowledgements
We are grateful to the Centre for eResearch, the University of Auckland, for access to computing resources such as research virtual machines.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































