Finding genetic variants responsible for human disease hiding in the universe of benign variants
Klaus Lehnert and Russell Snell, School of Biological Sciences
Our programs aim to unravel the genetic basis of human diseases using new approaches enabled by recent step-changes in genetic sequencing technologies (aka the “$1000 genome”).
The human genome comprises 3 billion loci and individuals typically differ from this ‘reference’ at millions of sites. These differences are the result of a complex interplay between ancient mutations, selection for survival fitness, mating between populations, events of near-extinction, and a very strong population expansion in the last 100 years.
A constant supply of new mutations creates new variants that are extremely rare. Some of these variants are directly responsible for disease and others cause genetic diseases in unknown combinations. We combine classical genetics approaches with genome sequencing to identify potentially disease-causing variants for experimental validation. One of our focus areas are neurological diseases, and we have just started a large project to understand the genetic nature of autism-spectrum disorders (www.mindsforminds.org.nz) – a debilitating neurodevelopmental condition with increasing prevalence in all human populations. We expect that identification of genetic mutations will help us to better understand the disease process and identify new targets for therapeutic intervention. This is ‘big data’ research – we typically process 100 billion ‘data points’ for each family, and the data analysis and storage requirements have posed significant challenges to traditionally data-poor biomedical analysis.
Analysing human genomes
Using the parallel processing options available on the Pan cluster we were able to derive optimal combinations for multiple interdependent parameters to align 100’s of millions of sequence reads to the human genome. These read sequences are strings of 100 nucleotide ‘characters’ (one of the four DNA ‘bases’ plus ‘not known’), including a confidence score for each base call, and contain a small number of differences to the reference string – some of these are the patient’s individual variants, and some are technical errors.
The non-uniform nature of the human genome reference further complicates the alignment problem. The goal is to assign a unique position for each read in the genome using mixed algorithms employing string matching to ‘seed’ the alignment and a combination of string matching and similarity scoring to extend the alignment through gaps and differences taking into account the confidence score for each base call. Through parameter optimisation and multi-threading we successfully reduced run times from several hours to seven minutes per patient.
The second step of each patient’s genome analysis aims to derive ‘genotypes’ for each of the millions of loci that differ from the reference in each patient. A genotype is a ‘best call’ for the two characters that can be observed at a single position (humans have two non-identical copies of each gene, one inherited from each parent). Genome sequencing creates 20-500 individual ‘observations’ of each position and the observations may be inconsistent, and/or may be different to the reference. We obtain a ‘consensus call’ for each position through a process that first proposes a de novo solution for the variant locus (i.e., not influenced by the reference), and then applies complex Bayesian framework to compute the most probable genotype at each locus.
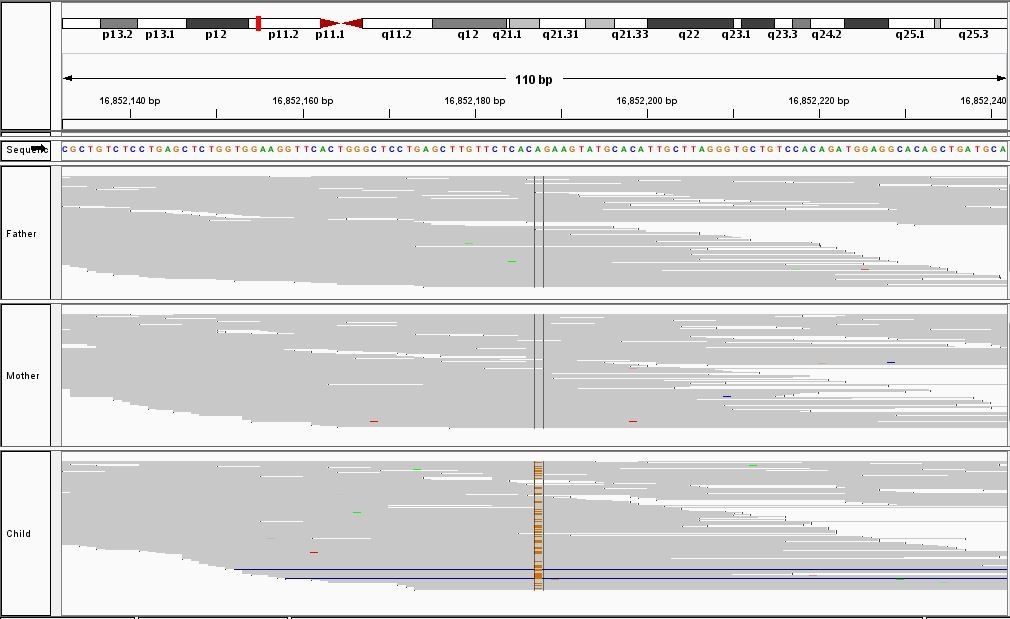
This process requires approximately 2000 hours of computation for a single family. However, on the Pan cluster we can apply a classic scatter-gather approach: we split the genome into dozens of segments, compute genotypes and probabilities for all segments in parallel, then combine the results from the individual computations to generate a list of all variants in each individual – with confidence scores. Total compute time remains unchanged, but the process completes overnight! We use standard bioinformatics software programs such as Burrows-Wheeler aligner, the Genome Analysis Toolkit (GATK), samtools, and others; these work very well but require multi-dimensional parameter optimisation. Figure 1 is an example of the alignments we work with.
It shows a software visualisation of the alignment of 360 million 100-nucleotide reads for a parent-child trio against an unrelated genome reference. Our analysis on Pan has clearly identified a new mutation in the child (orange squares, bottom panel), and it appears to affect only one of the child’s two gene copies. Being able to obtain results in hours instead of months will not only permit us to analyse more patients than otherwise possible, it also makes it possible to evaluate the effects of reduced coverage on genotyping confidence. If successful, this will reduce our cost of data generation, which in turn will increase the number of cases we can analyse.
What next?
While our program is under way, we are now starting collaboration with other researchers investigating other genetic diseases around New Zealand. The Pan cluster is the perfect home for this collaboration, and we hope to enrich further optimisation of our approaches. And of course there are disease-causing variants to be discovered!