
Genomic Virtual Lab (GVL) as a bioinformatics training platform
Peter Tsai, PhD Candidate, Dr Thierry Jean Lints, Professional Teaching Fellow, Professor Cristin Print, Molecular Medicine and Pathology; Dr William Schierding, Research Fellow, Liggins Institute
Introduction
The ability to analyse large-scale next-generation sequencing data is now an essential skill in modern biology. There is an increasing demand within the Faculty of Medical Health Sciences at the University in bioinformatics training to better understand how to analyse RNA (ribonucleic acid)* – Sequencing data, rather than using out-of-shelf end-to-end black box solutions.

The workshop
The week-long RNA-Sequencing workshop initiated by Professor Cristin Print from Faculty of Medical and Health Sciences, with a team effort from William Schierding, Peter Tsai and Thierry Lints, was held on the 19th February this year. It aims to provide an intensive step-by-step training to help researchers to understand how sequencing data needs to be analysed. The workshop on the analysis of RNA-sequencing data will include hands-on analysis of a real dataset, taking participants through a complete analysis pipeline from raw sequencing data to output graphics and spreadsheets, it covers:
- Sequencing platforms and terminology
- Gene expression and transcriptomics
- Exploring public data sources
- Pathway analysis

Genomics Virtual Laboratory (GVL)**
With the significant inputs from Sean Matheny from Centre for eResearch, the workshop uses the GVL training platform running on NeCTAR***.
GVL is a web portal provides a growing suite of genomics analysis tools, the biologists can start working immediately with no setup required. Date can be analysed easily on the NeCTAR research cloud. A workbench for reproducible results, a command line interface to support bioinformaticians, and tutorials to help biologists learn analysis techniques.
It is normally time consuming and sometimes difficult process to set up a bioinformatics analysis system. The pre-configured GVL eliminated the need to set up the system from scratch. The demand for the workshop amongst researchers has been overwhelming, and another weeklong workshop has been planned for the end of November 2018 for those researchers who could not attend the first workshop. We hope the workshop will provide a good understanding of the tools and processes used, and enable researchers to analyse simple RNA-sequencing datasets themselves.

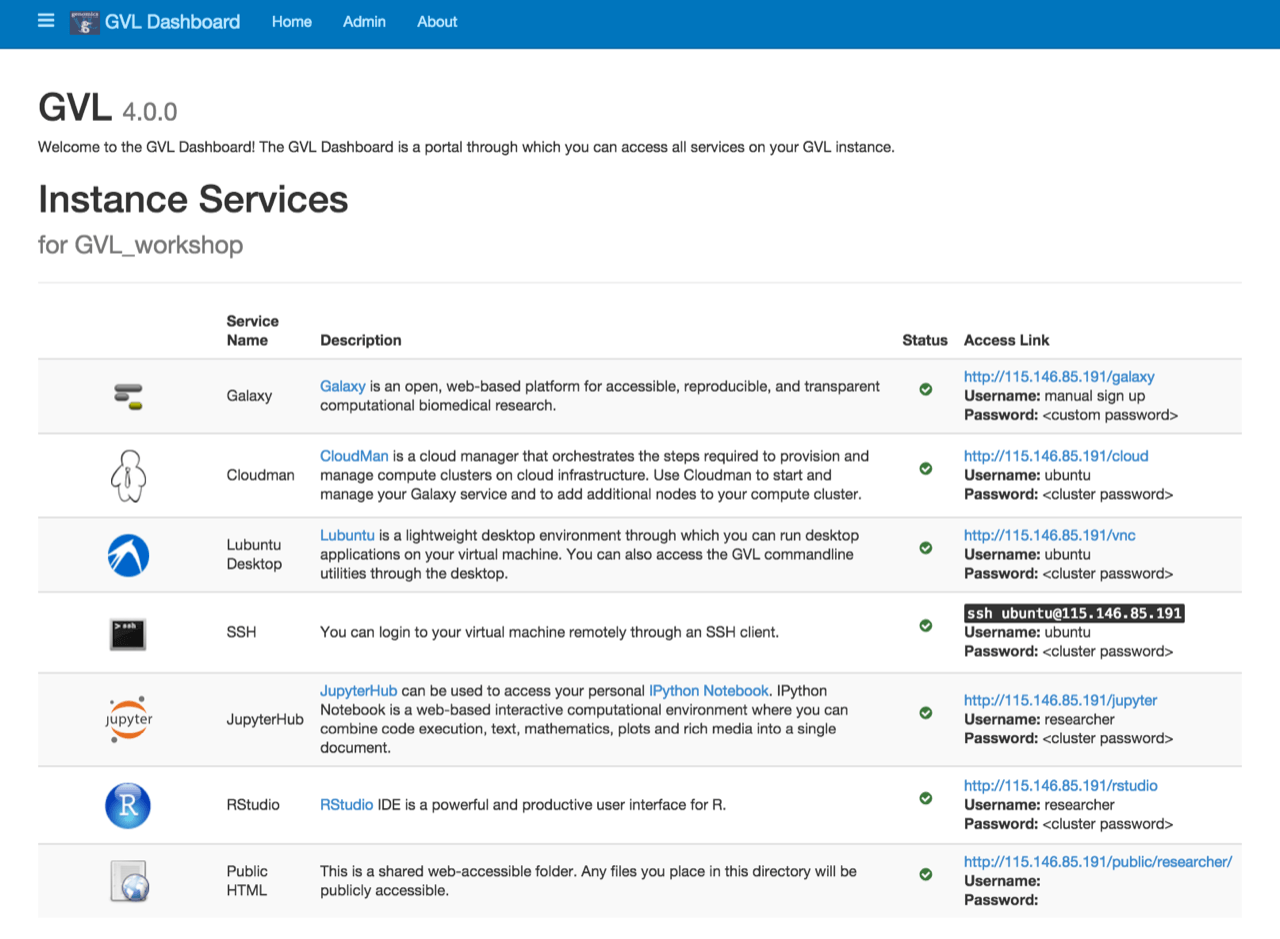
Image source: A screenshot of the GVL dashboard which lists various instance services it can run from NeCTAR.
Reference:
* RNA: Ribonucleic acid, a nucleic acid present in all living cells. Its principal role is to act as a messenger carrying instructions from DNA for controlling the synthesis of proteins, although in some viruses RNA rather than DNA carries the genetic information.
** GVL development has been led by the Victorian Life Sciences Computation Initiative at the University of Melbourne in collaboration with the Research Computing Centre at the University of Queensland, and acknowledges funding from the NeCTAR project.
*** NeCTAR: The National eResearch Collaboration Tools and Resources project provides an online infrastructure that supports researchers to connect with colleagues in Australia and around the world. The University of Auckland has joined NeCTAR in 2017 and can now easily add New Zealand collaborators who log in via the REANNZ Tuakiri service. Improved access to the Nectar Cloud for NZ researchers supports the Science, Research and Innovation Cooperation Agreement signed by Australia and New Zealand in early 2017.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































