
Mathematically modelling gastrointestinal electrical activity
Shameer Sathar, Jerry Gao, Mark Trew, Leo Cheng, Auckland Bioengineering Institute
The gastrointestinal system
The gastrointestinal (GI) system comprises a series of discrete organs, from mouth to anus, with distinct functions – digestion, absorption of nutrients, excretion, and protection from digestive agents and pathogens.
GI motility, the regulated contraction of muscles along the GI tract, is coordinated by the underlying omnipresent electrical activity termed as slow waves. These are initiated and propagated through networks of the interstitial cells of Cajal (ICC) to the smooth muscle cells (SMC).
Gastric dysrhythmias, or abnormal electrical activity in the stomach, have been implicated in the pathophysiology of several motility disorders, such as gastroparesis. A validated mathematical model offers a virtual medium in which a variety of hypotheses can be comprehensively investigated. The effects of different treatment strategies can also be predicted, potentially more cost effectively than by other experimental approaches.
An exciting application, is using models to relate body surface electric field, or magnetic field, potentials to the underlying gastric slow wave activity, as shown in Figure 1. Modelling on the Pan cluster
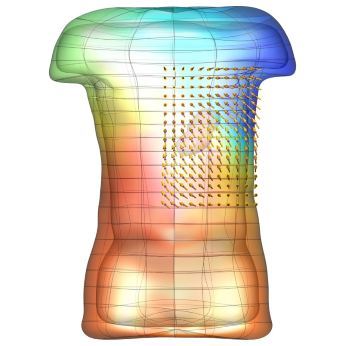
Figure 1: Torso model with electric potential fields (coloured surface) and magnetic fields (gold arrows) resulting from simulated gastric slow wave activity. Work conducted in conjunction with Prof. Alan Bradshaw (Vanderbilt University, Nashville, TN).
Modelling on the Pan cluster
Multiscale models of gastric electrophysiology (encompassing the subcellular, cell, tissue, organ, and whole-body scales) can be mathematically described by continuum modelling frameworks such as the monodomain or bidomain equations. Spatial discretisation of the 3D geometrical domain, using a finite element method, leads to a system of differential-algebraic equations. A semi-implicit method is used for temporal discretisation, where the linear term is treated implicitly and the nonlinear terms explicitly. The numerical solution of these equations on tissue-specific geometric models is typically computationally expensive. The CHASTE computational package, originally developed by the Computational Biology Group at the University of Oxford for solving tissue electrophysiology problems with cardiac applications, is being used for this research. It is built upon an MPI-based library, enabling it to efficiently split the high computational demands of simulation over many parallel processes. The CHASTE code scales linearly across multiple cores of the NeSI Pan cluster.
Recently, we developed a mathematical model for quantifying ICC network-function relationships by embedding biophysically-based ICC cell models into tissue-specific ICC network imaging data. Figure 2 shows the activation-time map of slow waves over an ICC network. This particular problem was relatively small, with about 300,000 solution points and a simulation time of 1000 ms. However, multiple sets of ICC network samples were analysed in parallel, as multiple independent jobs, each using up to 32 cores. The entire set of ICC networks were fully analysed in less than 48 hours using the cluster. If the NeSI Pan cluster had not been available, this data may have required weeks for analysis.
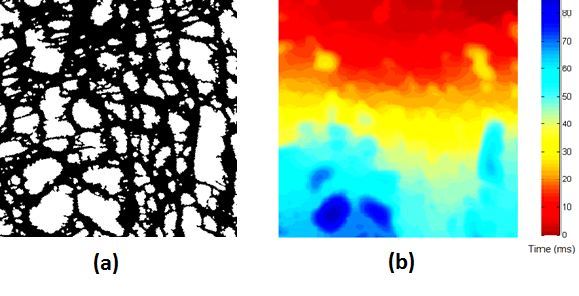
Figure 2. Slow wave simulation over an ICC network. (a) Tissue-specific ICC network imaging data. (b) Activation time map of slow waves over the network.

Figure 3. Simulated slow wave activity on an anatomically realistic stomach geometry.
What next?
In the future, we plan to simulate gastric electrophysiology on a 3D computational domain (approximately 3 million computational nodes) over temporal durations of up to several minutes. This model will incorporate recently developed, biophysically based, ICC and SMC cell models, along with analysed ICC network function data. We aim to simultaneously utilise up to 1024 cores of the NeSI Pan cluster and concurrently employ the cluster GPU resources. Figure 3 shows preliminary results for slow wave simulations in the stomach.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































