
Modelling dispersal and ecological competition in a statistical phylogeographic framework
Louis Ranjard, David Welch, Marie Paturel and Stéphane Guindon, Department of Statistics
Developing a new model for reconstructing colonisation history of habitats
Competition between organisms influences the processes governing the colonisation of new habitats. As a consequence, species or populations arriving first at a suitable location may prevent secondary colonisation. While adaptation to environmental variables (e.g., temperature, altitude, etc.) is essential, the presence or absence of certain species at a particular location often depends on whether or not competing species co-occur. Despite the potential of competition to maintain populations in isolation, past quantitative analyses of evolution have largely ignored that factor because of technical difficulties.
We have developed a model that integrates competition along with dispersal into a Bayesian phylogeographic framework. The proposed model takes genetic sequences and their spatial coordinates as input in order to date dispersal events and estimate dispersal and competition parameters.
Simulating landscape colonisation
Our statistical modelling on Pan uses the R software and high throughput is obtained by using parallel processing. In the first step to get an accurate understanding of how our approach performs, an extensive test on the NeSI Pan cluster was done by (i) performing simulations of organisms colonising space and competing while evolving and (ii) estimating the competitive and dispersal parameters from the genetic diversity generated by these simulations (Figure 1).
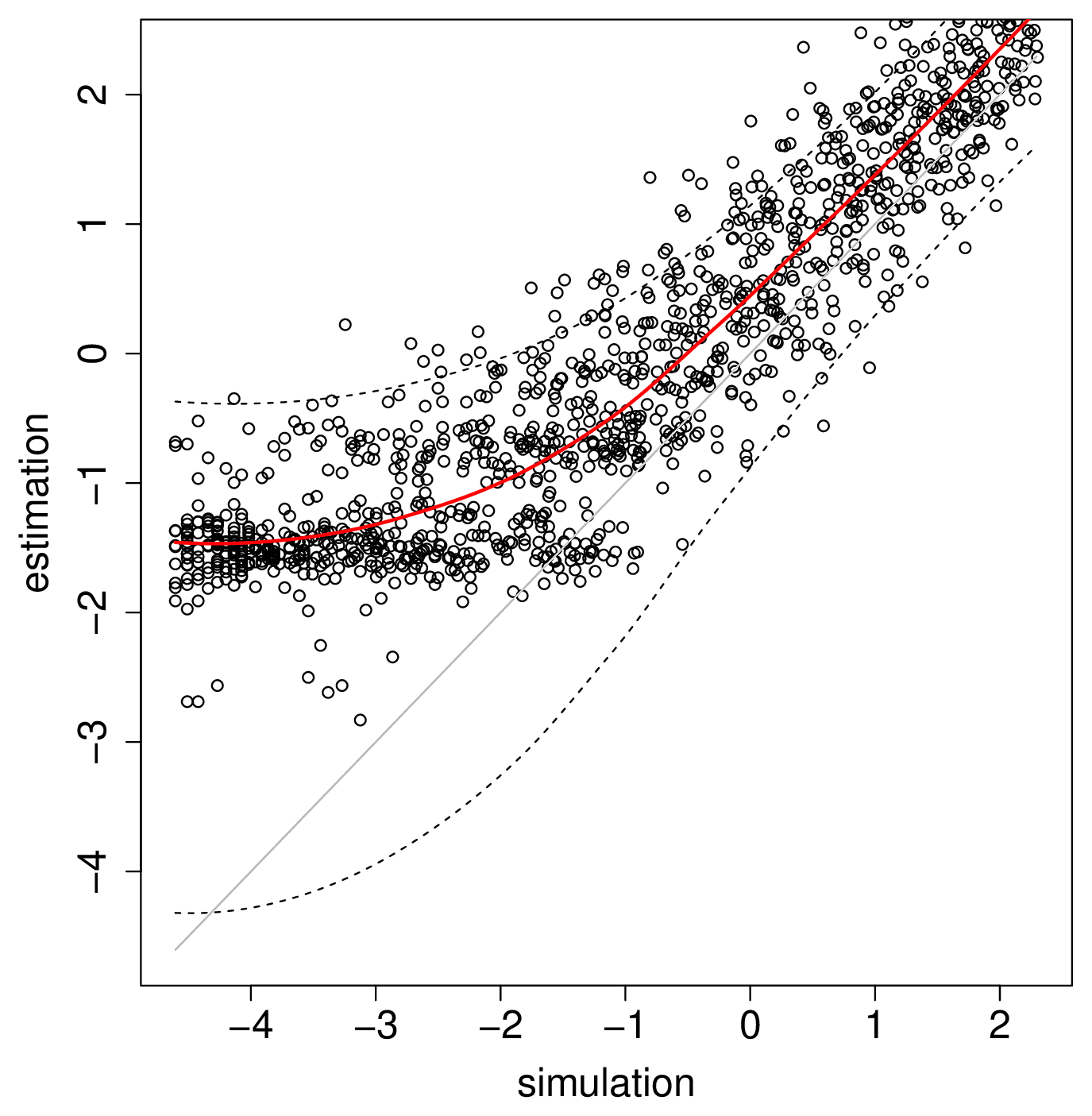
Figure 1: Results from simulations shows that our approach can successfully detect the influence of competition on the history of colonisation of space inferred from molecular sequences. Each dot on the graph corresponds to a different simulation instance performed on the Pan cluster.
In the second step, Pan was used to analyse a dataset of Hawaiian endemic species of the Banza genus called Hawaiian katydids –grasshopper-like insects. The genetic relationships among these organisms show evidence for island radiation with different species inhabiting the Hawaiian archipelago.
The phylogenetic tree built from mitochondrial sequences exhibits a striking “progression rule” pattern whereby each island appears to have been colonised only once. No obvious environmental feature varying across islands explains the observed geographical distribution of these organisms. Therefore, adaptation does not seem to be the factor causing the absence of secondary colonisaton.
We fitted our model to a rooted phylogenetic tree with node heights expressed in calendar time units (Figure 2). Using Pan we could demonstrate that competition can be detected with high sensitivity and specificity from the analysis of genetic variations in space.
In the next step, we are planning on extending our model to integrate extra environmental parameters that may have affected the colonisation of space. The NeSI Pan cluster will be used similarly to investigate the characteristics of the new model and how it can be applied to any biogeographical datasets for which genetic data is available.
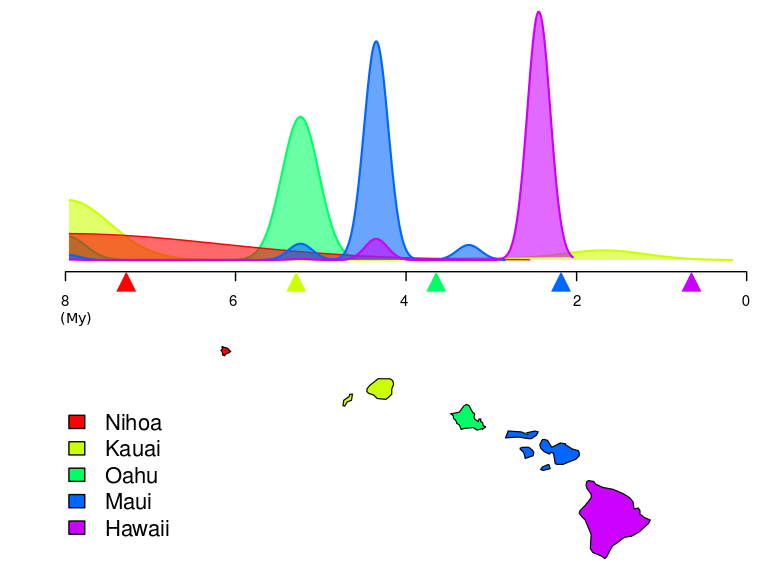
Figure 2: Colonisation date estimates of each Hawaiian Island from molecular sequences for the Hawaiian katydids (in million years, My). Geological estimates of islands formation are indicated by coloured triangles under the time axis. While the order of colonisation and geological formation agree, the time shift between the two dating approaches can probably be explained by an underestimate of the genetic mutation rate in these organisms.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































