
Multiscale modelling of saliva secretion
James Sneyd, Department of Mathematics, University of Auckland; David Yule, School of Medicine and Dentistry, University of Rochester; John Rugis, New Zealand eScience Infrastructure
Introduction
This interdisciplinary project encompasses a range of activities targeting anatomical data based structural modelling of individual salivary cell clusters, solution of cellular calcium dynamics function in full 3D simulations, interactive visualisation of resultant calcium waves and validation of results by comparison to experimental data. The model will be used to test duct cell function and for the testing of pathological conditions. The overall project is funded by the National Institutes of Health, USA (Sneyd, Yule).
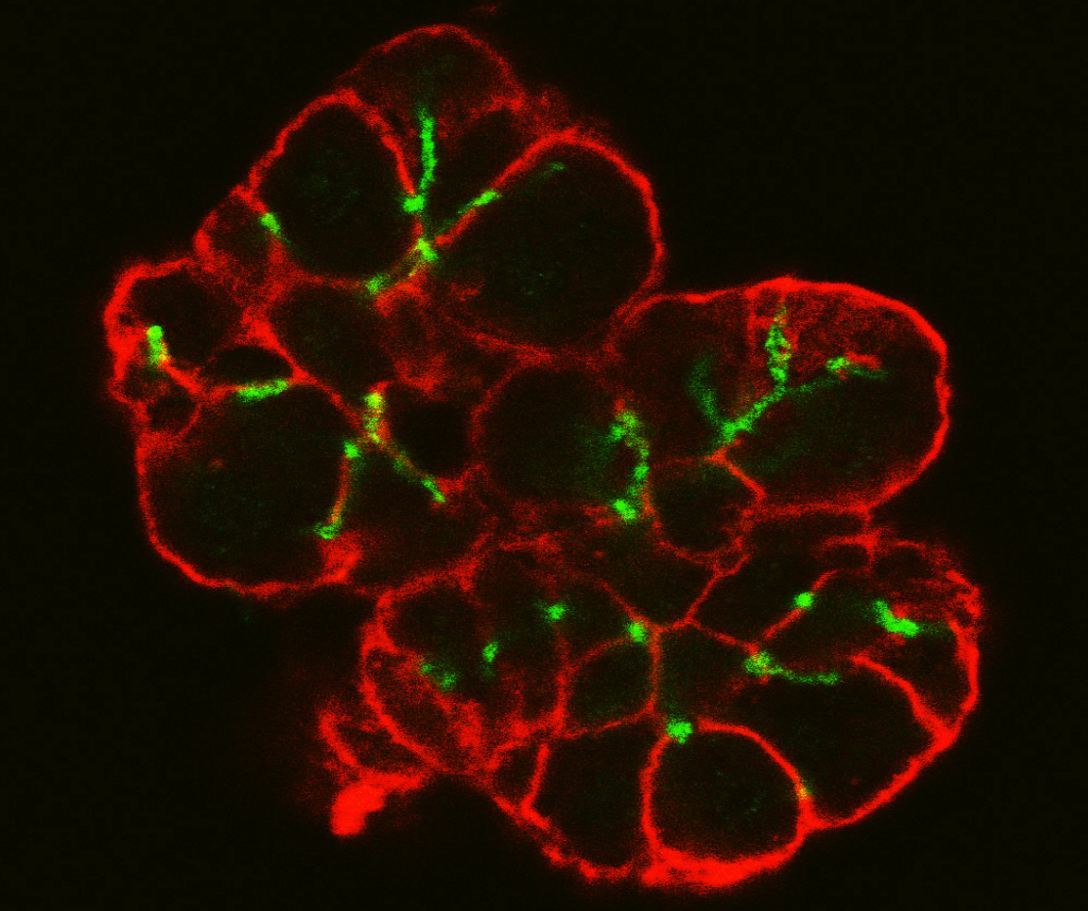
Figure 1: Colour coded digitised image slice.
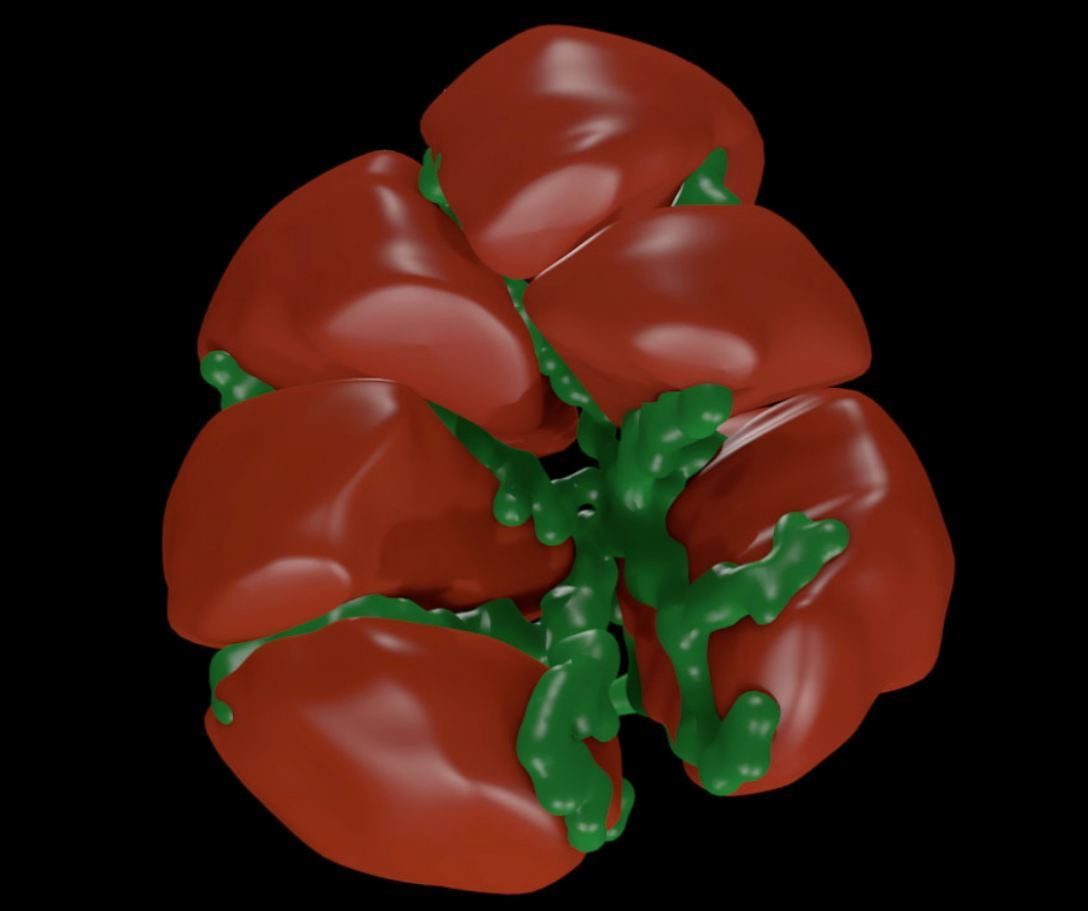
Figure 2: Full 3D mesh model of a cluster of cells.
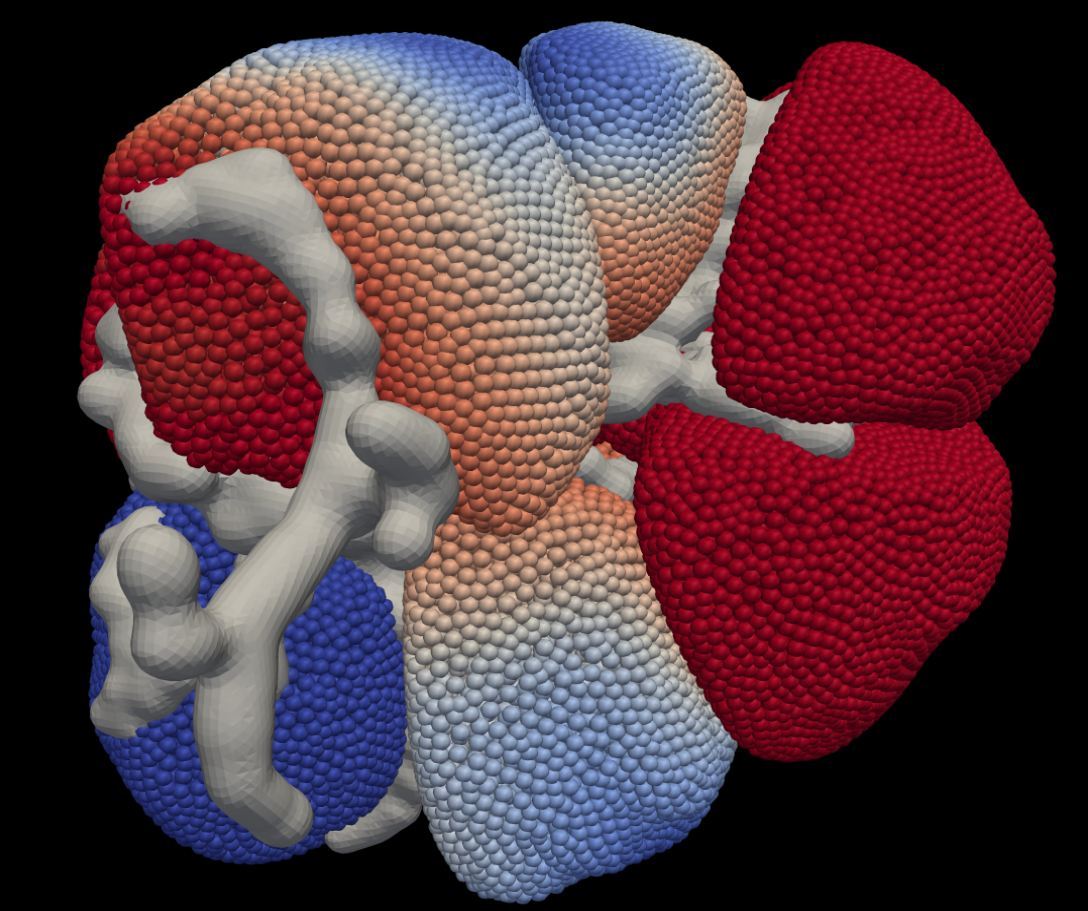
Figure 3: Simulation output snapshot.
Current activity and results
Real biological samples where digitized using fluorescent markers and confocal microscopy. A sample image slice in which individual cell outlines can be seen is shown in Figure 1. The cell membranes are colour coded red and the interconnecting lumen is colour coded green. Note that, in living beings, the saliva secreted from the cells is transported through the assumed tube-like lumen structure. The full set of images slices was used as the basis for a full 3D graphics model reconstruction of one cluster of cells as shown in Figure 2. The tube-like structure of the lumen can now be clearly seen. This anatomically correct model was used in turn as the basis for the creation of a 3D tetrahedral mesh suitable for finite element simulations.
The same underlying 3D graphics mesh was used in the animated visualisation of the calcium concentration simulation time series results. One time series frame is shown in Figure 3.Through the NeSI Pan cluster was used for both graphics model rendering and running the finite element simulations. Thanks to the NeSI, we were able to render higher quality images and run many more simulation variants than would have been possible on a desktop computer. This facility will also enable us to scale up our model to include many more cells.
What’s next
As expected, simulation results for each of the cells differ somewhat. Further work will include a detailed analysis of how cell geometry effects the generation and propagation of calcium waves within each cell. We also plan to construct a larger model based on new digitisations using refined microscopy techniques.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































