
Phylogeny and phylogeography of the family kyphosidae (Perciformes: teleostei)
Steen Wilhelm Knudsen, School of Biological Sciences
Introduction
Comparing DNA sequences of multiple genes in various species is commonly applied today for inferring relationships among species and finding the best phylogenetic tree that explains how species have evolved and how they are related.
In this study we focused on the sea chubs (family Kyphosidae) that are abundant consumers of macroalgae on both temperate and tropical reef systems. Several species have extremely broad distributions. Juveniles of Kyphosus species are often found well offshore among drift algae, suggesting a high potential for oceanic dispersal. The relationships and taxonomic status of the 16 valid species of sea chubs (genera Hermosilla, Kyphosus, Neoscorpis and Sectator) known worldwide have long been problematical due to perceived lack of character differentiation, complicating ecological assessment.
We inferred the relationship and distribution of all known species of sea chubs using a combined analysis of partial DNA fragments from mitochondrial markers (12s, 16s, cytb, tRNA -Pro, -Phe, -Thr and -Val) and three nuclear markers (rag1, rag2, tmo4c4), in total comprising 5960 bp from 118 individuals. Synonyms among Atlantic and Indo-Pacific taxa show that several sea chub species are more widespread than previously thought.

Figure 1: A photo of Kyphosus gladius (above) and Kyphosus sydneyanus (below). We discovered that Kyphosus gladius had gone unnoticed in Western Australia, as it has been confused K. sydneyanus in the past. But careful examination of radiograph photographs and detailed analysis of the DNA sequence variation in the different species of Kyphosus lead us to conclude that K. gladius should be described as a new species for Western Australia. These findings were published last year in Zootaxa (vol. 3599). Photo by K. Clements.
Using the Pan cluster
The main work carried out on the Pan cluster comprised parallel analysis using the software MrBayes and BEAST, and was run across multiple CPUs where each result afterwards was combined and the best tree inferred through the lengthy analysis that occasionally can take more than a day to complete. Furthermore, by applying fossil calibrations on the groups of species in the tree while the analysis runs, the time for speciation of different species and groups can be inferred and provide a chronogram – an evolutionary tree that shows points in time of speciation for various groups.
The chronogram shows that Kyphosus originated relatively recently, in the early Miocene. Neoscorpis lithophilus, Kyphosus cornelii and Hermosilla azurea are basal in the topology, implying a subtropical, Indo-Pacific origin for the family. The Southern Indo-Pacific Ocean hosts the greatest diversity in sea chubs, and Western Australia and South-Eastern Africa appear to be refuge habitats for ancestral lineages. The tropical species of Kyphosus and Sectator represent very recent divergences, indicating that herbivory in the group originated at higher latitudes, and that carnivory in the autapomorphic Sectator ocyurus is derived. Several clades of perciform herbivorous reef fishes had established themselves on tropical reefs in the Eocene. Kyphosids colonised low latitude reefs much more recently, and speciated rapidly.
Running these analyses on the Pan cluster made it possible to infer evolutionary trees that are based on more comprehensive data analysis than would be possible – within a feasible timeframe – to prepare from analysis run on a single desktop computer. It also made it possible to test multiple settings and cross influence of parameters to ensure the resulting trees were the most optimal reflection of the evolution among the sea chubs that could be inferred from the inherent DNA data. Being able to infer these evolutionary trees through the use of parallel computing on Pan cluster was fundamental to this study, and ensured that the evolutionary scenario inferred was based on rigorous testing of the underlying data. Our research has resulted in two scientific publications in the journal Zootaxa, and we are currently working on two additional scientific papers, and two chapters for a guidebook on New Zealand fishes.
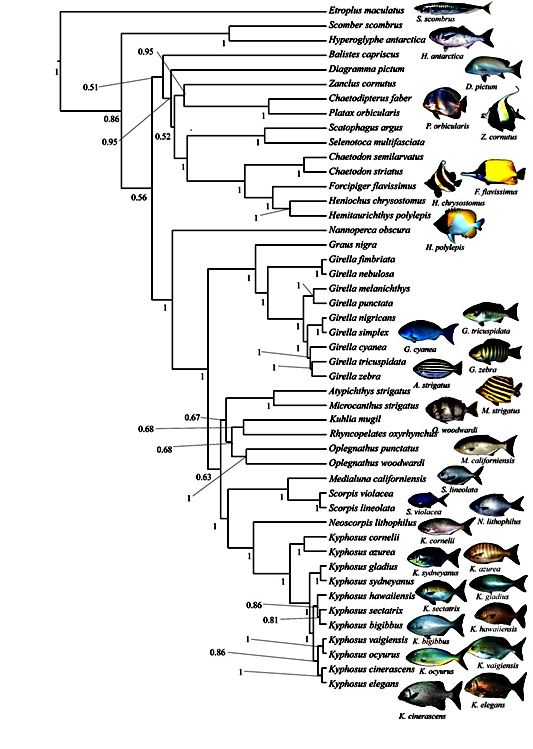
Figure 2: Evolutionary tree based on 5578 bp from both mtDNA (3427 bp) and nDNA (899 bp rag1, 772 bp rag2 and 479 bp tmo4c4), and inferred in BEAST by running the analysis 160 mio. generations, in 8 parallel runs, sampling a total of 10,000 trees and creating a consensus tree from 8,000 trees. The consensus tree shows that the genera Scorpis, Medialuna and Neoscorpis and Kyphosus are closely related.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































