
Using GPUs to expand our understanding of the Solar System
Dr Philip Sharp, Department of Mathmatics
Using High Performance Computing (HPC) to study the early evolution of the Solar System
Dr Sharp is collaborating with William Newman, a Professor in Physics & Astronomy at UCLA, to investigate whether the Nice model, the leading model for the Solar System’s evolution, accurately predicts the present day orbits of the giant planets, Jupiter, Saturn, Uranus and Neptune. In addition to the giant planets, the Nice model also includes the Sun and N-5 small bodies called planetesimals (see Figure 1).
“Put simply, the work would not be possible without HPC,” Sharp says. “We chose NeSI because its HPC resources are the best in the country.”
As part of his investigation, Sharp uses mathematical methods called N-body simulations. His latest simulation program, running on a single Graphics Processing Unit (GPU), has performed simulations of 20 to 100 million years with N=1024 astronomical bodies. Although this value of N has been used by other research groups, Sharp and Newman are the first to perform accurate simulations with this value. Results of that work were published in the Journal of Computational Science.
“Collisionless N-body simulations over tens of millions of years are an important tool in understanding the early evolution of planetary systems,” says Sharp. “Our method is significantly more accurate than symplectic methods and sufficiently fast.”
So far, results from these simulations show the Nice model is unstable for the number of bodies researchers use with the model. This instability therefore limits its predictive power. To strengthen this argument, Sharp and his colleagues wanted to perform accurate simulations with larger values of N, however they were limited by their existing simulation program because the elapsed time would be too large.

Figure 1. Initial configuration of a simulation (not to scale), showing the Sun (gold), large planets (red) and planetesimals (silver).
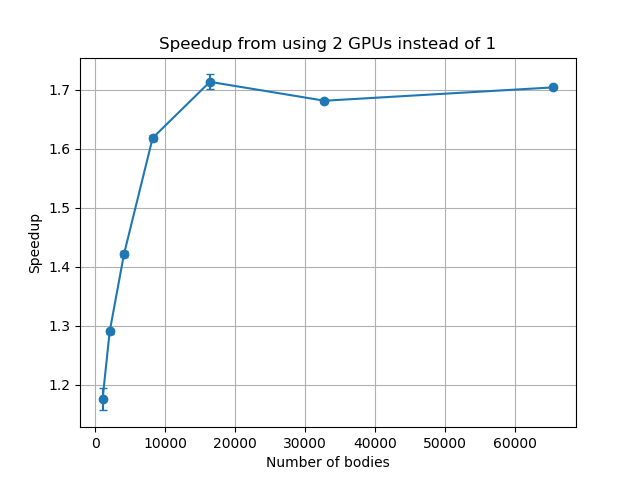
Figure 2. The speedups obtained by running on two GPUs compared to one. The initial increase is due to the GPUs not being fully utilised at lower numbers of bodies.
The help of NeSI’ s Scientific Programmer
This is where Chris Scott from NeSI’s Computational Science Team stepped in to help. Although Sharp’s simulation program was already well optimised, further gains could be made by modifying the program to run on multiple GPUs at once. With Scott’s help, the researchers were able to convert their simulation program to make use of all available GPUs within a node from a single CPU core. They applied this to a range of numbers of bodies, from 1,024 to 65,532, and immediately observed a modest improvement in performance when working with the lower numbers of bodies. As they switched to larger numbers of bodies, which are of more interest to Sharp, the gains were even greater, with a 1.7x speedup observed for simulations with 16,384 or more bodies. Scott then experimented with introducing OpenMP, an Application Programming Interface (API) commonly used for parallel programming, and the combined speedups from using two GPUs and OpenMP were 1.9x for 4,096 bodies, 2.0x for 16,384 bodies, and 1.8x for 32,768 bodies. As a result of these changes, the simulation program is now running up to two times faster than previously,” says Scott. “Also, these gains will allow researchers to run simulations of larger systems in a more reasonable time and verify the results obtained from their calculations.” “Tackling these types of research questions relies heavily on access to High Performance Computing,” says Sharp.
As they continue their work in this area, the insights uncovered by Sharp and his colleagues will further strengthen the Nice model as a tool for explaining how the Solar System is evolving. “Our results clearly signpost what to do next – perform simulations with a lot more bodies,” Sharp says. “We expect, as happens with galactic simulations, that increasing the number of bodies will make the Nice model more stable and hence improve its predictive power.”
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































