
Using research virtual machines to analyse fMRI datasets
Dr Reece Roberts, Research Fellow, Psychology
Introduction
Magnetic resonance imaging (MRI) allows clinicians and researchers to leverage differences in the magnetic properties of different tissue types and/or blood to generate images of brain structure and function (fMRI). A number of research groups at The University of Auckland, in both the Faculties of Science and Medical and Health Sciences collect MRI data of various types at the Centre for Advanced Magnetic Resonance Imaging (CAMRI; https://www.fmhs.auckland.ac.nz/en/faculty/camri.html). In the case of fMRI, these data are 4D datasets comprised of a series of 3D images (each acquired every 1-3 seconds). A number of research groups have collaborated with CeR to build virtual machines suited to analysing these large, complex datasets.
Data organisation
An increasingly common way to structure data is to use the Brain Imaging Data Structure format (BIDS; http://bids.neuroimaging.io/), which is a movement to standardise data structures across the worldwide neuroimaging community, to allow for greater sharing of data and an increase in reproducibility of neuroimaging research.
Data quality assurance and preprocessing
Adopting the BIDS data structure allow for the use of a number of open-source packages to be applied to a given dataset. For example, automated quality assurance of MRI images (https://github.com/poldracklab/mriqc) produces metrics enabling the detection of outliers on a range of measures (see Fig. 1).
To enable the analysis of group effects, MRI scans from study participants are preprocessed (using, e.g., fMRIprep; https://github.com/poldracklab/fmriprep) to remove sources of variance that are unrelated to the signal of interest (e.g. motion and scanner artefacts, physiological noise). In addition, the images of each individual are normalised to be in a common space. Performing these analyses on CeR VMs has required the installation of Docker packages, and CeR staff have been critical in getting these up-and-running. This is particularly helpful as Docker containers are not currently supported on NeSI HOC platform.
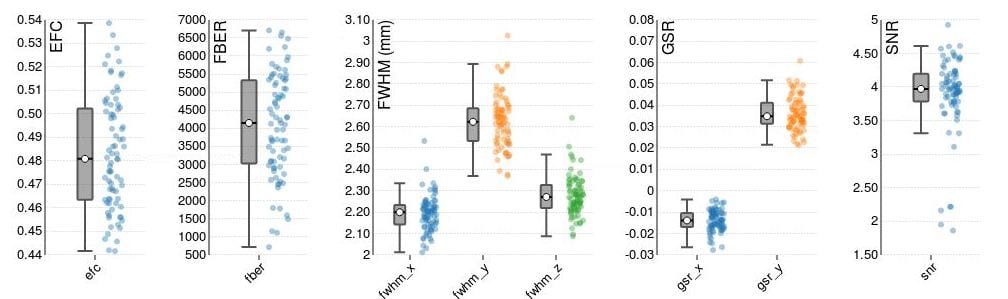
Fig. 1. Some example parameters from automatic quality assurance of MRI images that allow for automatic rejection of outliers.
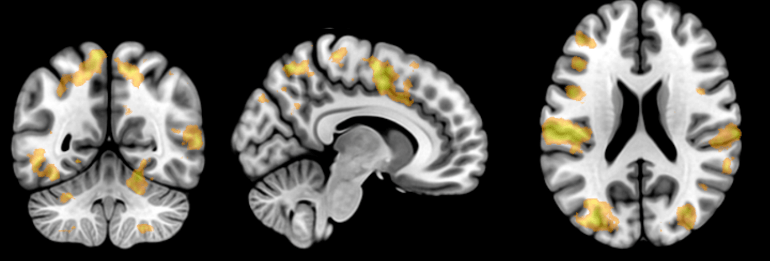
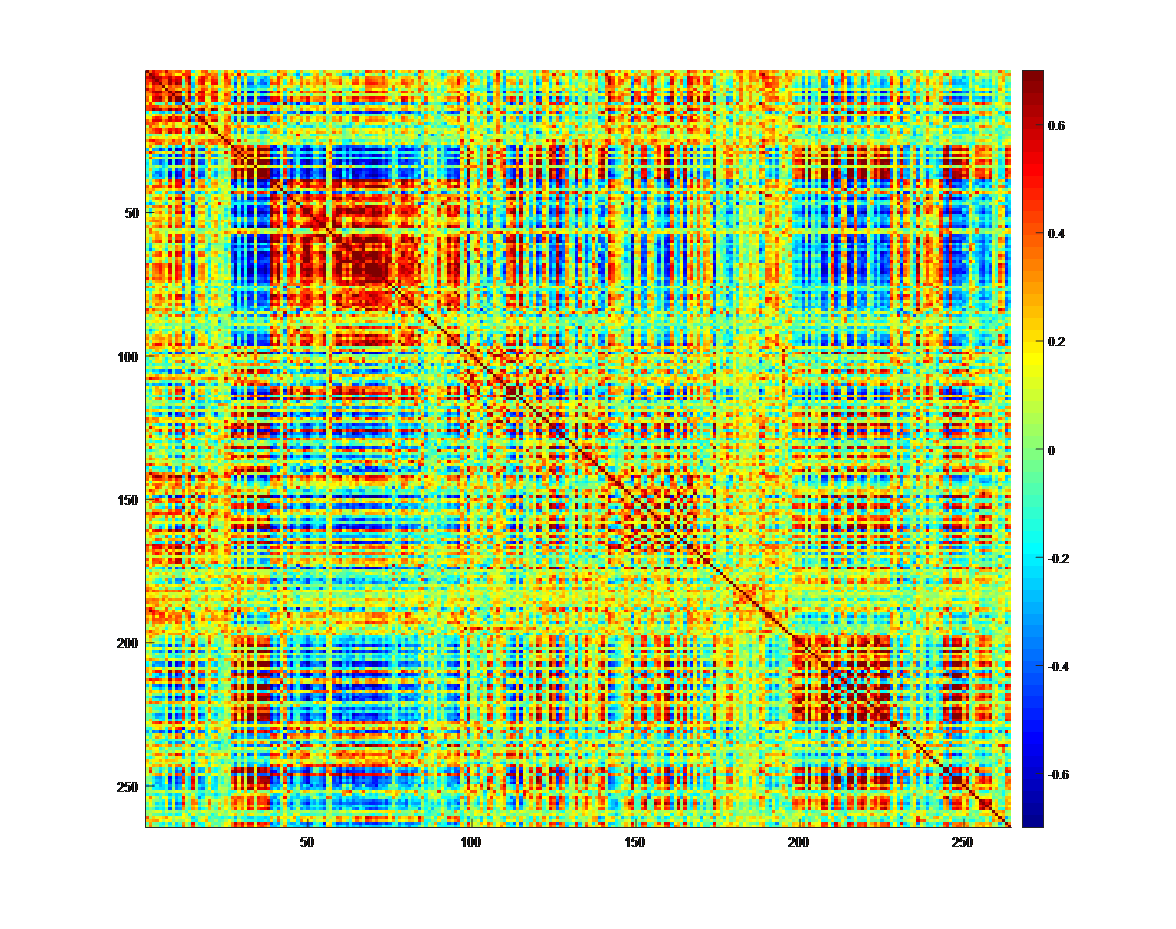
Fig. 3. On the top, a statistical parametric map showing widespread increases in brain activity in response to a cognitive task. At the bottom, a correlation matrix of 264 regions of interest (ROIs) showing the strength of the temporal correlation between the time-series
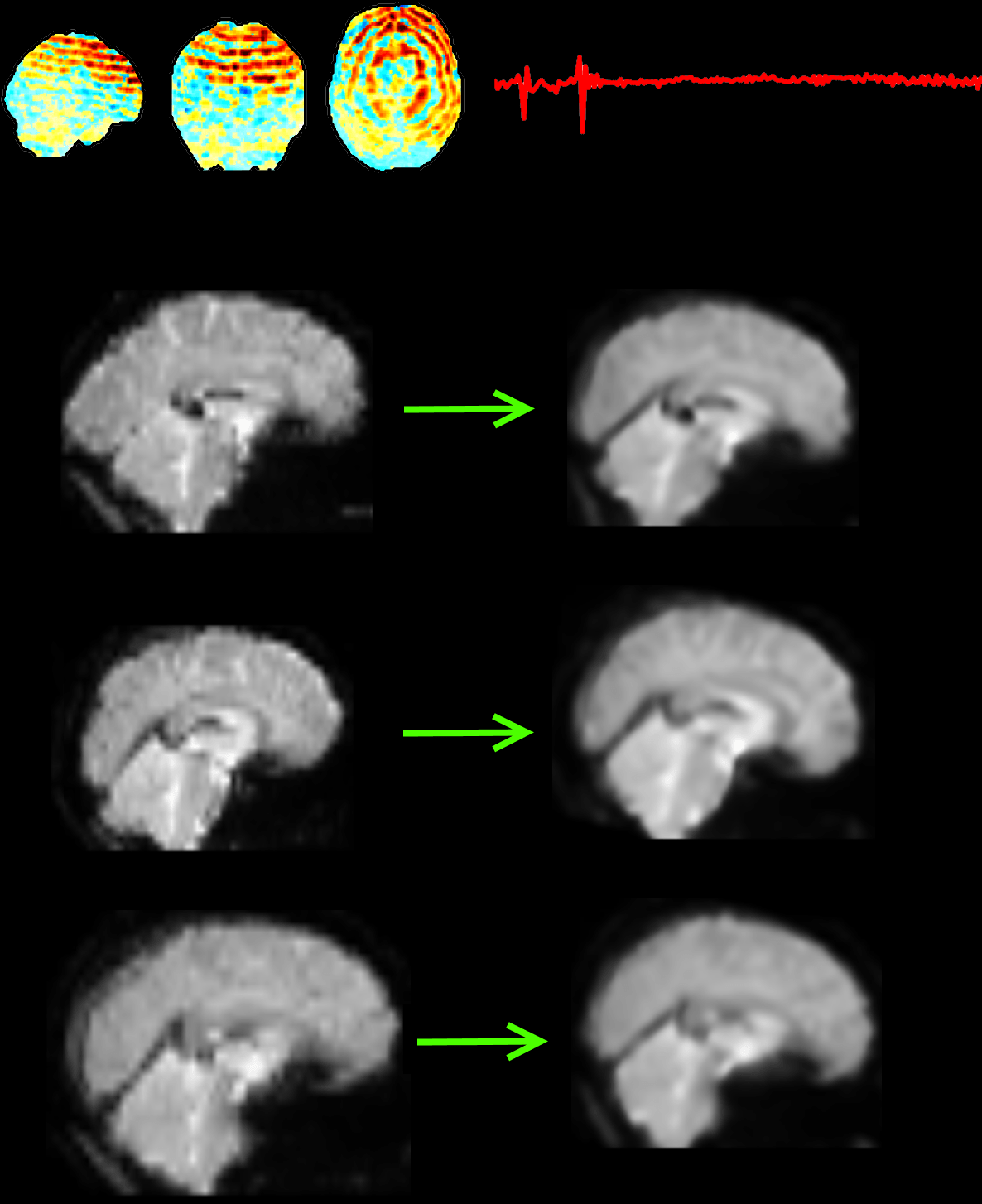
Fig. 2. The top row shows an example of a motion artefact component in fMRI data. Below are three participants with different shaped brains. The images from each participant are normalised to a common template, allowing for group analyses to be performed.
See more case study projects

Our Voices: using innovative techniques to collect, analyse and amplify the lived experiences of young people in Aotearoa

Painting the brain: multiplexed tissue labelling of human brain tissue to facilitate discoveries in neuroanatomy

Detecting anomalous matches in professional sports: a novel approach using advanced anomaly detection techniques

Benefits of linking routine medical records to the GUiNZ longitudinal birth cohort: Childhood injury predictors

Using a virtual machine-based machine learning algorithm to obtain comprehensive behavioural information in an in vivo Alzheimer’s disease model

Mapping livability: the “15-minute city” concept for car-dependent districts in Auckland, New Zealand

Travelling Heads – Measuring Reproducibility and Repeatability of Magnetic Resonance Imaging in Dementia

Novel Subject-Specific Method of Visualising Group Differences from Multiple DTI Metrics without Averaging

Re-assess urban spaces under COVID-19 impact: sensing Auckland social ‘hotspots’ with mobile location data

Aotearoa New Zealand’s changing coastline – Resilience to Nature’s Challenges (National Science Challenge)

Proteins under a computational microscope: designing in-silico strategies to understand and develop molecular functionalities in Life Sciences and Engineering

Coastal image classification and nalysis based on convolutional neural betworks and pattern recognition

Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis

Measuring impact of entrepreneurship activities on students’ mindset, capabilities and entrepreneurial intentions

Using Zebra Finch data and deep learning classification to identify individual bird calls from audio recordings

Automated measurement of intracranial cerebrospinal fluid volume and outcome after endovascular thrombectomy for ischemic stroke

Using simple models to explore complex dynamics: A case study of macomona liliana (wedge-shell) and nutrient variations

Fully coupled thermo-hydro-mechanical modelling of permeability enhancement by the finite element method

Modelling dual reflux pressure swing adsorption (DR-PSA) units for gas separation in natural gas processing

Molecular phylogenetics uses genetic data to reconstruct the evolutionary history of individuals, populations or species

Wandering around the molecular landscape: embracing virtual reality as a research showcasing outreach and teaching tool
























































































































































